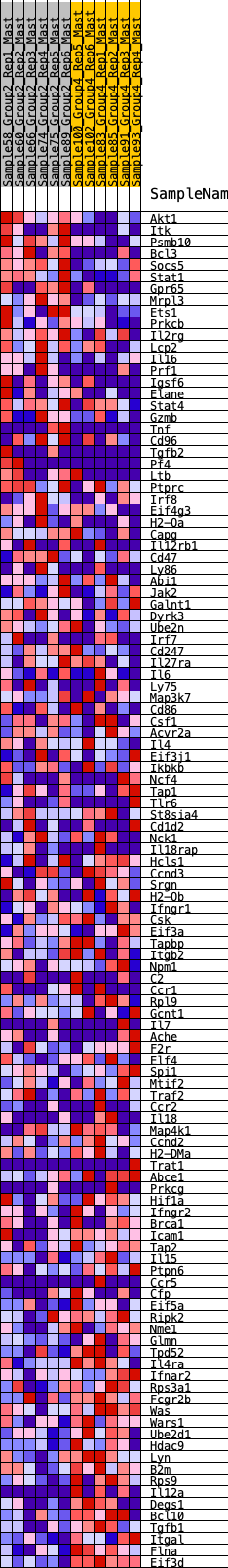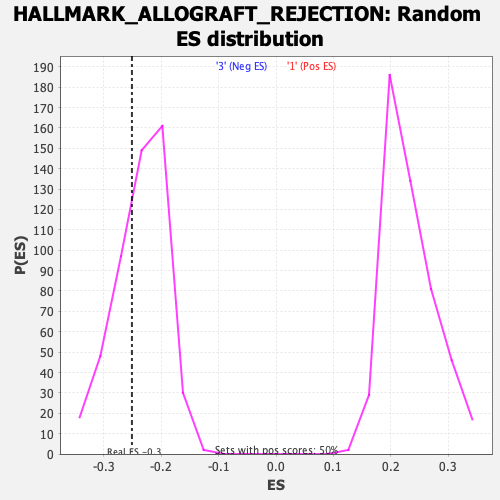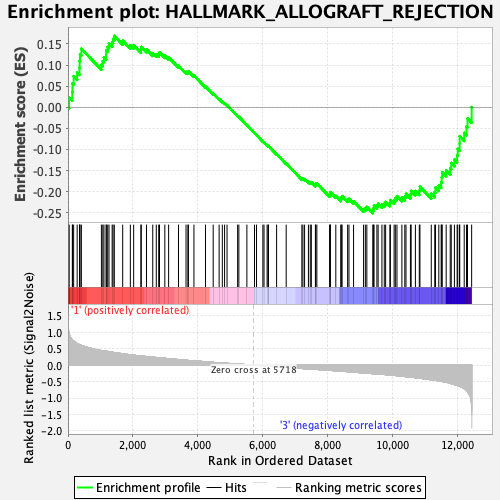
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.2511001 |
| Normalized Enrichment Score (NES) | -1.0696812 |
| Nominal p-value | 0.32475248 |
| FDR q-value | 0.7367349 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 36 | 0.968 | 0.0232 | No |
| 2 | Itk | 133 | 0.773 | 0.0362 | No |
| 3 | Psmb10 | 139 | 0.766 | 0.0565 | No |
| 4 | Bcl3 | 174 | 0.732 | 0.0735 | No |
| 5 | Socs5 | 282 | 0.654 | 0.0824 | No |
| 6 | Stat1 | 354 | 0.625 | 0.0935 | No |
| 7 | Gpr65 | 359 | 0.623 | 0.1100 | No |
| 8 | Mrpl3 | 371 | 0.618 | 0.1258 | No |
| 9 | Ets1 | 412 | 0.604 | 0.1388 | No |
| 10 | Prkcb | 1029 | 0.443 | 0.1008 | No |
| 11 | Il2rg | 1072 | 0.436 | 0.1092 | No |
| 12 | Lcp2 | 1107 | 0.432 | 0.1181 | No |
| 13 | Il16 | 1177 | 0.424 | 0.1239 | No |
| 14 | Prf1 | 1178 | 0.424 | 0.1353 | No |
| 15 | Igsf6 | 1223 | 0.415 | 0.1430 | No |
| 16 | Elane | 1263 | 0.408 | 0.1508 | No |
| 17 | Stat4 | 1360 | 0.394 | 0.1536 | No |
| 18 | Gzmb | 1388 | 0.389 | 0.1620 | No |
| 19 | Tnf | 1427 | 0.383 | 0.1692 | No |
| 20 | Cd96 | 1685 | 0.348 | 0.1577 | No |
| 21 | Tgfb2 | 1921 | 0.314 | 0.1472 | No |
| 22 | Pf4 | 2023 | 0.304 | 0.1471 | No |
| 23 | Ltb | 2246 | 0.277 | 0.1366 | No |
| 24 | Ptprc | 2257 | 0.276 | 0.1432 | No |
| 25 | Irf8 | 2418 | 0.261 | 0.1373 | No |
| 26 | Eif4g3 | 2611 | 0.242 | 0.1282 | No |
| 27 | H2-Oa | 2720 | 0.232 | 0.1257 | No |
| 28 | Capg | 2795 | 0.226 | 0.1258 | No |
| 29 | Il12rb1 | 2819 | 0.224 | 0.1300 | No |
| 30 | Cd47 | 2983 | 0.209 | 0.1224 | No |
| 31 | Ly86 | 3100 | 0.197 | 0.1183 | No |
| 32 | Abi1 | 3404 | 0.171 | 0.0984 | No |
| 33 | Jak2 | 3636 | 0.150 | 0.0837 | No |
| 34 | Galnt1 | 3694 | 0.144 | 0.0829 | No |
| 35 | Dyrk3 | 3710 | 0.143 | 0.0855 | No |
| 36 | Ube2n | 3880 | 0.130 | 0.0754 | No |
| 37 | Irf7 | 4236 | 0.104 | 0.0493 | No |
| 38 | Cd247 | 4473 | 0.084 | 0.0325 | No |
| 39 | Il27ra | 4656 | 0.070 | 0.0196 | No |
| 40 | Il6 | 4754 | 0.064 | 0.0135 | No |
| 41 | Ly75 | 4824 | 0.060 | 0.0095 | No |
| 42 | Map3k7 | 4900 | 0.055 | 0.0049 | No |
| 43 | Cd86 | 5226 | 0.035 | -0.0205 | No |
| 44 | Csf1 | 5264 | 0.032 | -0.0227 | No |
| 45 | Acvr2a | 5511 | 0.016 | -0.0422 | No |
| 46 | Il4 | 5746 | -0.000 | -0.0612 | No |
| 47 | Eif3j1 | 5808 | -0.004 | -0.0660 | No |
| 48 | Ikbkb | 6003 | -0.018 | -0.0813 | No |
| 49 | Ncf4 | 6039 | -0.022 | -0.0835 | No |
| 50 | Tap1 | 6134 | -0.028 | -0.0904 | No |
| 51 | Tlr6 | 6164 | -0.030 | -0.0919 | No |
| 52 | St8sia4 | 6177 | -0.031 | -0.0921 | No |
| 53 | Cd1d2 | 6427 | -0.049 | -0.1109 | No |
| 54 | Nck1 | 6722 | -0.069 | -0.1329 | No |
| 55 | Il18rap | 7205 | -0.101 | -0.1693 | No |
| 56 | Hcls1 | 7238 | -0.103 | -0.1691 | No |
| 57 | Ccnd3 | 7289 | -0.106 | -0.1703 | No |
| 58 | Srgn | 7407 | -0.114 | -0.1767 | No |
| 59 | H2-Ob | 7464 | -0.117 | -0.1781 | No |
| 60 | Ifngr1 | 7497 | -0.119 | -0.1775 | No |
| 61 | Csk | 7622 | -0.128 | -0.1841 | No |
| 62 | Eif3a | 7632 | -0.128 | -0.1814 | No |
| 63 | Tapbp | 7667 | -0.131 | -0.1806 | No |
| 64 | Itgb2 | 8059 | -0.157 | -0.2081 | No |
| 65 | Npm1 | 8081 | -0.159 | -0.2055 | No |
| 66 | C2 | 8082 | -0.159 | -0.2012 | No |
| 67 | Ccr1 | 8251 | -0.171 | -0.2102 | No |
| 68 | Rpl9 | 8403 | -0.183 | -0.2175 | No |
| 69 | Gcnt1 | 8421 | -0.184 | -0.2140 | No |
| 70 | Il7 | 8445 | -0.185 | -0.2108 | No |
| 71 | Ache | 8615 | -0.197 | -0.2192 | No |
| 72 | F2r | 8656 | -0.200 | -0.2171 | No |
| 73 | Elf4 | 8798 | -0.211 | -0.2228 | No |
| 74 | Spi1 | 9106 | -0.236 | -0.2414 | No |
| 75 | Mtif2 | 9161 | -0.241 | -0.2393 | No |
| 76 | Traf2 | 9207 | -0.244 | -0.2363 | No |
| 77 | Ccr2 | 9390 | -0.257 | -0.2442 | Yes |
| 78 | Il18 | 9416 | -0.259 | -0.2392 | Yes |
| 79 | Map4k1 | 9427 | -0.260 | -0.2330 | Yes |
| 80 | Ccnd2 | 9517 | -0.267 | -0.2330 | Yes |
| 81 | H2-DMa | 9555 | -0.269 | -0.2288 | Yes |
| 82 | Trat1 | 9672 | -0.278 | -0.2307 | Yes |
| 83 | Abce1 | 9744 | -0.284 | -0.2288 | Yes |
| 84 | Prkcg | 9786 | -0.287 | -0.2244 | Yes |
| 85 | Hif1a | 9917 | -0.299 | -0.2268 | Yes |
| 86 | Ifngr2 | 9933 | -0.301 | -0.2199 | Yes |
| 87 | Brca1 | 10047 | -0.311 | -0.2207 | Yes |
| 88 | Icam1 | 10085 | -0.314 | -0.2153 | Yes |
| 89 | Tap2 | 10135 | -0.319 | -0.2106 | Yes |
| 90 | Il15 | 10287 | -0.335 | -0.2138 | Yes |
| 91 | Ptpn6 | 10377 | -0.345 | -0.2118 | Yes |
| 92 | Ccr5 | 10413 | -0.349 | -0.2052 | Yes |
| 93 | Cfp | 10550 | -0.363 | -0.2064 | Yes |
| 94 | Eif5a | 10572 | -0.366 | -0.1983 | Yes |
| 95 | Ripk2 | 10703 | -0.381 | -0.1985 | Yes |
| 96 | Nme1 | 10823 | -0.394 | -0.1976 | Yes |
| 97 | Glmn | 10843 | -0.396 | -0.1884 | Yes |
| 98 | Tpd52 | 11192 | -0.443 | -0.2047 | Yes |
| 99 | Il4ra | 11300 | -0.459 | -0.2010 | Yes |
| 100 | Ifnar2 | 11323 | -0.463 | -0.1903 | Yes |
| 101 | Rps3a1 | 11424 | -0.479 | -0.1855 | Yes |
| 102 | Fcgr2b | 11497 | -0.492 | -0.1781 | Yes |
| 103 | Was | 11514 | -0.493 | -0.1661 | Yes |
| 104 | Wars1 | 11528 | -0.496 | -0.1538 | Yes |
| 105 | Ube2d1 | 11651 | -0.521 | -0.1496 | Yes |
| 106 | Hdac9 | 11776 | -0.553 | -0.1448 | Yes |
| 107 | Lyn | 11807 | -0.562 | -0.1320 | Yes |
| 108 | B2m | 11905 | -0.592 | -0.1239 | Yes |
| 109 | Rps9 | 11989 | -0.618 | -0.1140 | Yes |
| 110 | Il12a | 12008 | -0.623 | -0.0987 | Yes |
| 111 | Degs1 | 12065 | -0.640 | -0.0860 | Yes |
| 112 | Bcl10 | 12067 | -0.641 | -0.0688 | Yes |
| 113 | Tgfb1 | 12207 | -0.714 | -0.0608 | Yes |
| 114 | Itgal | 12278 | -0.771 | -0.0457 | Yes |
| 115 | Flna | 12308 | -0.803 | -0.0264 | Yes |
| 116 | Eif3d | 12436 | -1.375 | 0.0004 | Yes |