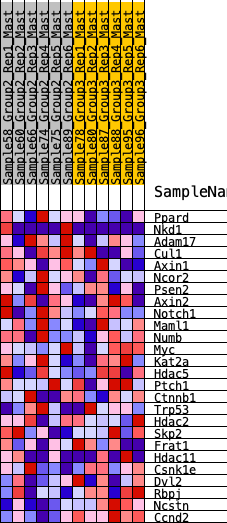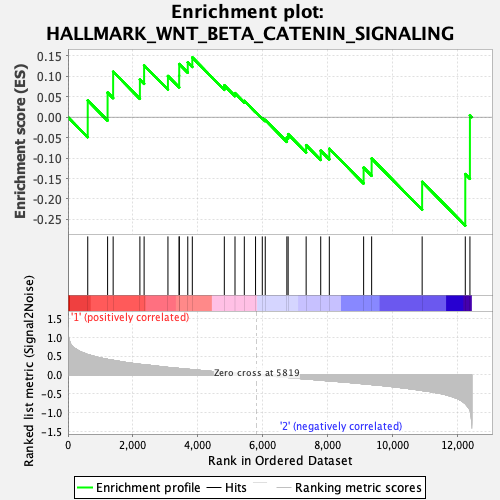
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.26510864 |
| Normalized Enrichment Score (NES) | -0.84182024 |
| Nominal p-value | 0.68916154 |
| FDR q-value | 0.90067077 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ppard | 608 | 0.539 | 0.0408 | No |
| 2 | Nkd1 | 1220 | 0.412 | 0.0601 | No |
| 3 | Adam17 | 1391 | 0.388 | 0.1111 | No |
| 4 | Cul1 | 2215 | 0.282 | 0.0918 | No |
| 5 | Axin1 | 2345 | 0.270 | 0.1265 | No |
| 6 | Ncor2 | 3079 | 0.199 | 0.1006 | No |
| 7 | Psen2 | 3425 | 0.172 | 0.1014 | No |
| 8 | Axin2 | 3427 | 0.172 | 0.1299 | No |
| 9 | Notch1 | 3690 | 0.150 | 0.1339 | No |
| 10 | Maml1 | 3830 | 0.140 | 0.1461 | No |
| 11 | Numb | 4816 | 0.066 | 0.0778 | No |
| 12 | Myc | 5145 | 0.044 | 0.0587 | No |
| 13 | Kat2a | 5432 | 0.025 | 0.0397 | No |
| 14 | Hdac5 | 5778 | 0.002 | 0.0123 | No |
| 15 | Ptch1 | 5986 | -0.011 | -0.0025 | No |
| 16 | Ctnnb1 | 6077 | -0.017 | -0.0068 | No |
| 17 | Trp53 | 6743 | -0.064 | -0.0497 | No |
| 18 | Hdac2 | 6782 | -0.067 | -0.0416 | No |
| 19 | Skp2 | 7337 | -0.106 | -0.0686 | No |
| 20 | Frat1 | 7786 | -0.136 | -0.0819 | No |
| 21 | Hdac11 | 8051 | -0.156 | -0.0772 | No |
| 22 | Csnk1e | 9108 | -0.233 | -0.1235 | No |
| 23 | Dvl2 | 9355 | -0.253 | -0.1011 | No |
| 24 | Rbpj | 10911 | -0.409 | -0.1581 | Yes |
| 25 | Ncstn | 12240 | -0.756 | -0.1392 | Yes |
| 26 | Ccnd2 | 12383 | -0.932 | 0.0047 | Yes |