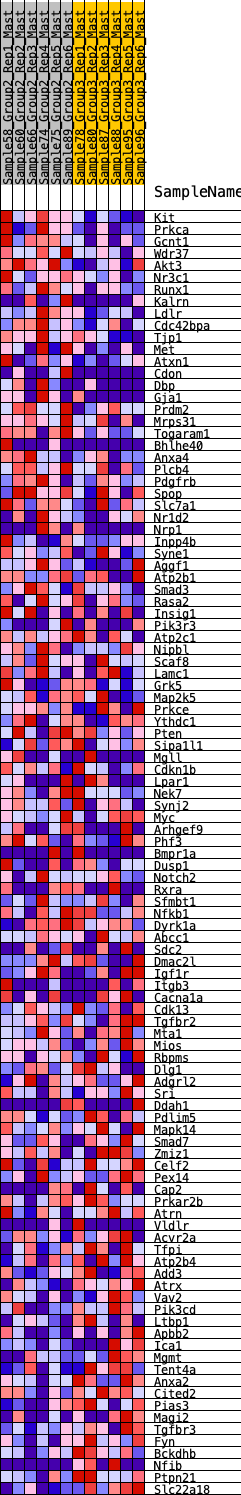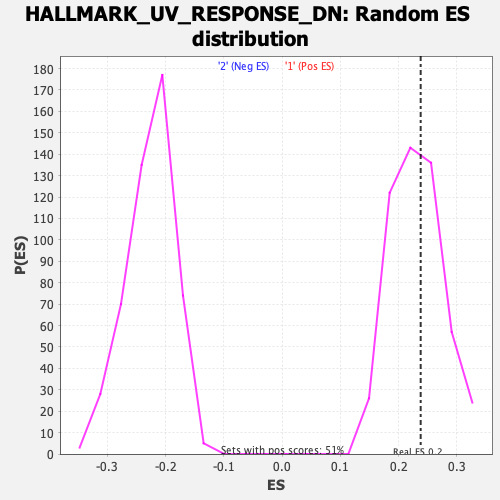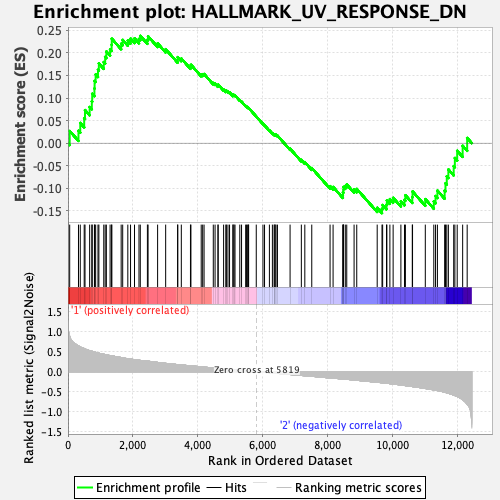
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.23767133 |
| Normalized Enrichment Score (NES) | 1.0330614 |
| Nominal p-value | 0.42716536 |
| FDR q-value | 0.8987406 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Kit | 52 | 0.890 | 0.0269 | Yes |
| 2 | Prkca | 323 | 0.643 | 0.0276 | Yes |
| 3 | Gcnt1 | 378 | 0.617 | 0.0448 | Yes |
| 4 | Wdr37 | 499 | 0.579 | 0.0553 | Yes |
| 5 | Akt3 | 524 | 0.567 | 0.0733 | Yes |
| 6 | Nr3c1 | 667 | 0.526 | 0.0802 | Yes |
| 7 | Runx1 | 734 | 0.509 | 0.0926 | Yes |
| 8 | Kalrn | 746 | 0.507 | 0.1095 | Yes |
| 9 | Ldlr | 814 | 0.489 | 0.1212 | Yes |
| 10 | Cdc42bpa | 820 | 0.488 | 0.1379 | Yes |
| 11 | Tjp1 | 852 | 0.480 | 0.1522 | Yes |
| 12 | Met | 924 | 0.465 | 0.1627 | Yes |
| 13 | Atxn1 | 951 | 0.460 | 0.1767 | Yes |
| 14 | Cdon | 1102 | 0.434 | 0.1797 | Yes |
| 15 | Dbp | 1145 | 0.426 | 0.1912 | Yes |
| 16 | Gja1 | 1182 | 0.421 | 0.2031 | Yes |
| 17 | Prdm2 | 1297 | 0.402 | 0.2079 | Yes |
| 18 | Mrps31 | 1344 | 0.394 | 0.2179 | Yes |
| 19 | Togaram1 | 1349 | 0.393 | 0.2314 | Yes |
| 20 | Bhlhe40 | 1638 | 0.352 | 0.2204 | Yes |
| 21 | Anxa4 | 1685 | 0.344 | 0.2287 | Yes |
| 22 | Plcb4 | 1843 | 0.323 | 0.2273 | Yes |
| 23 | Pdgfrb | 1927 | 0.313 | 0.2315 | Yes |
| 24 | Spop | 2049 | 0.299 | 0.2322 | Yes |
| 25 | Slc7a1 | 2187 | 0.284 | 0.2310 | Yes |
| 26 | Nr1d2 | 2227 | 0.280 | 0.2377 | Yes |
| 27 | Nrp1 | 2446 | 0.263 | 0.2292 | No |
| 28 | Inpp4b | 2467 | 0.260 | 0.2367 | No |
| 29 | Syne1 | 2761 | 0.230 | 0.2210 | No |
| 30 | Aggf1 | 3009 | 0.206 | 0.2082 | No |
| 31 | Atp2b1 | 3379 | 0.176 | 0.1844 | No |
| 32 | Smad3 | 3380 | 0.176 | 0.1906 | No |
| 33 | Rasa2 | 3491 | 0.166 | 0.1875 | No |
| 34 | Insig1 | 3773 | 0.144 | 0.1697 | No |
| 35 | Pik3r3 | 3787 | 0.143 | 0.1737 | No |
| 36 | Atp2c1 | 4104 | 0.118 | 0.1522 | No |
| 37 | Nipbl | 4149 | 0.116 | 0.1527 | No |
| 38 | Scaf8 | 4192 | 0.113 | 0.1532 | No |
| 39 | Lamc1 | 4476 | 0.089 | 0.1334 | No |
| 40 | Grk5 | 4531 | 0.084 | 0.1320 | No |
| 41 | Map2k5 | 4620 | 0.078 | 0.1276 | No |
| 42 | Prkce | 4626 | 0.077 | 0.1299 | No |
| 43 | Ythdc1 | 4794 | 0.068 | 0.1187 | No |
| 44 | Pten | 4863 | 0.063 | 0.1154 | No |
| 45 | Sipa1l1 | 4879 | 0.062 | 0.1164 | No |
| 46 | Mgll | 4928 | 0.059 | 0.1146 | No |
| 47 | Cdkn1b | 4977 | 0.056 | 0.1126 | No |
| 48 | Lpar1 | 5083 | 0.049 | 0.1058 | No |
| 49 | Nek7 | 5084 | 0.049 | 0.1075 | No |
| 50 | Synj2 | 5116 | 0.046 | 0.1066 | No |
| 51 | Myc | 5145 | 0.044 | 0.1059 | No |
| 52 | Arhgef9 | 5293 | 0.035 | 0.0952 | No |
| 53 | Phf3 | 5347 | 0.030 | 0.0920 | No |
| 54 | Bmpr1a | 5474 | 0.022 | 0.0825 | No |
| 55 | Dusp1 | 5501 | 0.021 | 0.0811 | No |
| 56 | Notch2 | 5535 | 0.018 | 0.0791 | No |
| 57 | Rxra | 5555 | 0.017 | 0.0782 | No |
| 58 | Sfmbt1 | 5565 | 0.016 | 0.0780 | No |
| 59 | Nfkb1 | 5800 | 0.002 | 0.0591 | No |
| 60 | Dyrk1a | 6006 | -0.012 | 0.0429 | No |
| 61 | Abcc1 | 6055 | -0.016 | 0.0396 | No |
| 62 | Sdc2 | 6205 | -0.026 | 0.0284 | No |
| 63 | Dmac2l | 6297 | -0.032 | 0.0221 | No |
| 64 | Igf1r | 6350 | -0.035 | 0.0192 | No |
| 65 | Itgb3 | 6373 | -0.037 | 0.0187 | No |
| 66 | Cacna1a | 6378 | -0.038 | 0.0197 | No |
| 67 | Cdk13 | 6409 | -0.041 | 0.0187 | No |
| 68 | Tgfbr2 | 6453 | -0.044 | 0.0167 | No |
| 69 | Mta1 | 6842 | -0.072 | -0.0122 | No |
| 70 | Mios | 7189 | -0.096 | -0.0369 | No |
| 71 | Rbpms | 7296 | -0.102 | -0.0419 | No |
| 72 | Dlg1 | 7510 | -0.117 | -0.0551 | No |
| 73 | Adgrl2 | 8074 | -0.158 | -0.0952 | No |
| 74 | Sri | 8167 | -0.164 | -0.0969 | No |
| 75 | Ddah1 | 8469 | -0.184 | -0.1149 | No |
| 76 | Pdlim5 | 8471 | -0.184 | -0.1086 | No |
| 77 | Mapk14 | 8487 | -0.185 | -0.1033 | No |
| 78 | Smad7 | 8489 | -0.185 | -0.0969 | No |
| 79 | Zmiz1 | 8543 | -0.188 | -0.0946 | No |
| 80 | Celf2 | 8588 | -0.192 | -0.0915 | No |
| 81 | Pex14 | 8814 | -0.211 | -0.1023 | No |
| 82 | Cap2 | 8896 | -0.216 | -0.1013 | No |
| 83 | Prkar2b | 9526 | -0.266 | -0.1430 | No |
| 84 | Atrn | 9674 | -0.278 | -0.1452 | No |
| 85 | Vldlr | 9694 | -0.280 | -0.1369 | No |
| 86 | Acvr2a | 9813 | -0.290 | -0.1363 | No |
| 87 | Tfpi | 9824 | -0.291 | -0.1269 | No |
| 88 | Atp2b4 | 9917 | -0.301 | -0.1239 | No |
| 89 | Add3 | 10017 | -0.310 | -0.1210 | No |
| 90 | Atrx | 10256 | -0.335 | -0.1286 | No |
| 91 | Vav2 | 10367 | -0.346 | -0.1254 | No |
| 92 | Pik3cd | 10395 | -0.350 | -0.1153 | No |
| 93 | Ltbp1 | 10607 | -0.373 | -0.1194 | No |
| 94 | Apbb2 | 10615 | -0.374 | -0.1069 | No |
| 95 | Ica1 | 11008 | -0.422 | -0.1239 | No |
| 96 | Mgmt | 11270 | -0.457 | -0.1290 | No |
| 97 | Tent4a | 11324 | -0.466 | -0.1170 | No |
| 98 | Anxa2 | 11381 | -0.474 | -0.1049 | No |
| 99 | Cited2 | 11604 | -0.514 | -0.1049 | No |
| 100 | Pias3 | 11630 | -0.520 | -0.0888 | No |
| 101 | Magi2 | 11670 | -0.531 | -0.0734 | No |
| 102 | Tgfbr3 | 11719 | -0.543 | -0.0582 | No |
| 103 | Fyn | 11883 | -0.588 | -0.0509 | No |
| 104 | Bckdhb | 11919 | -0.599 | -0.0328 | No |
| 105 | Nfib | 11991 | -0.621 | -0.0168 | No |
| 106 | Ptpn21 | 12159 | -0.699 | -0.0058 | No |
| 107 | Slc22a18 | 12298 | -0.817 | 0.0116 | No |