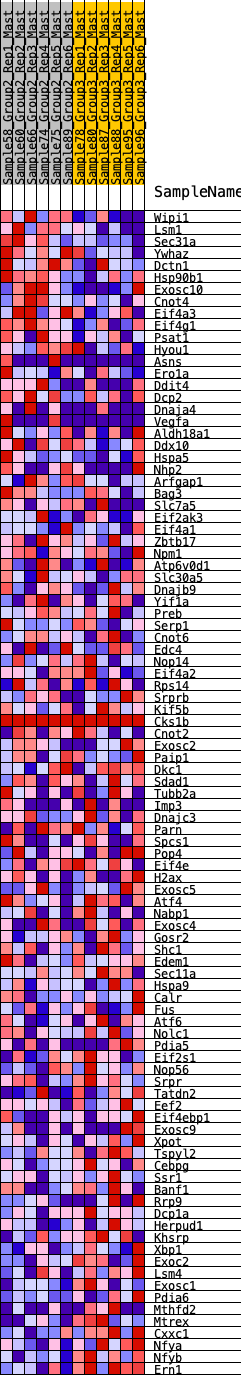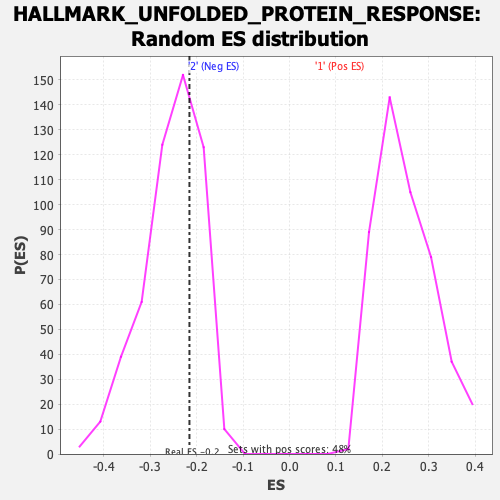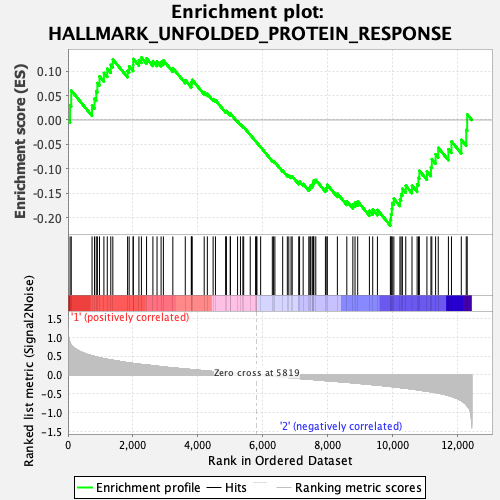
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.21579623 |
| Normalized Enrichment Score (NES) | -0.8493844 |
| Nominal p-value | 0.6857143 |
| FDR q-value | 0.9366817 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wipi1 | 64 | 0.861 | 0.0304 | No |
| 2 | Lsm1 | 100 | 0.792 | 0.0603 | No |
| 3 | Sec31a | 741 | 0.508 | 0.0294 | No |
| 4 | Ywhaz | 817 | 0.489 | 0.0435 | No |
| 5 | Dctn1 | 870 | 0.476 | 0.0590 | No |
| 6 | Hsp90b1 | 905 | 0.469 | 0.0756 | No |
| 7 | Exosc10 | 970 | 0.457 | 0.0893 | No |
| 8 | Cnot4 | 1106 | 0.433 | 0.0962 | No |
| 9 | Eif4a3 | 1210 | 0.415 | 0.1051 | No |
| 10 | Eif4g1 | 1318 | 0.398 | 0.1128 | No |
| 11 | Psat1 | 1381 | 0.390 | 0.1239 | No |
| 12 | Hyou1 | 1836 | 0.324 | 0.1005 | No |
| 13 | Asns | 1884 | 0.318 | 0.1099 | No |
| 14 | Ero1a | 2007 | 0.304 | 0.1125 | No |
| 15 | Ddit4 | 2010 | 0.304 | 0.1249 | No |
| 16 | Dcp2 | 2185 | 0.285 | 0.1226 | No |
| 17 | Dnaja4 | 2262 | 0.277 | 0.1279 | No |
| 18 | Vegfa | 2420 | 0.264 | 0.1261 | No |
| 19 | Aldh18a1 | 2616 | 0.245 | 0.1204 | No |
| 20 | Ddx10 | 2743 | 0.233 | 0.1198 | No |
| 21 | Hspa5 | 2868 | 0.219 | 0.1188 | No |
| 22 | Nhp2 | 2939 | 0.212 | 0.1219 | No |
| 23 | Arfgap1 | 3230 | 0.187 | 0.1061 | No |
| 24 | Bag3 | 3613 | 0.157 | 0.0816 | No |
| 25 | Slc7a5 | 3799 | 0.143 | 0.0725 | No |
| 26 | Eif2ak3 | 3804 | 0.142 | 0.0781 | No |
| 27 | Eif4a1 | 3827 | 0.141 | 0.0821 | No |
| 28 | Zbtb17 | 4196 | 0.112 | 0.0570 | No |
| 29 | Npm1 | 4293 | 0.105 | 0.0535 | No |
| 30 | Atp6v0d1 | 4471 | 0.090 | 0.0429 | No |
| 31 | Slc30a5 | 4543 | 0.083 | 0.0406 | No |
| 32 | Dnajb9 | 4856 | 0.064 | 0.0179 | No |
| 33 | Yif1a | 4881 | 0.062 | 0.0185 | No |
| 34 | Preb | 4999 | 0.054 | 0.0113 | No |
| 35 | Serp1 | 5004 | 0.054 | 0.0132 | No |
| 36 | Cnot6 | 5224 | 0.038 | -0.0030 | No |
| 37 | Edc4 | 5313 | 0.033 | -0.0087 | No |
| 38 | Nop14 | 5388 | 0.028 | -0.0136 | No |
| 39 | Eif4a2 | 5413 | 0.027 | -0.0144 | No |
| 40 | Rps14 | 5614 | 0.013 | -0.0300 | No |
| 41 | Srprb | 5779 | 0.002 | -0.0432 | No |
| 42 | Kif5b | 5805 | 0.001 | -0.0452 | No |
| 43 | Cks1b | 5821 | 0.000 | -0.0464 | No |
| 44 | Cnot2 | 5935 | -0.007 | -0.0553 | No |
| 45 | Exosc2 | 6295 | -0.032 | -0.0831 | No |
| 46 | Paip1 | 6334 | -0.034 | -0.0847 | No |
| 47 | Dkc1 | 6374 | -0.037 | -0.0863 | No |
| 48 | Sdad1 | 6614 | -0.055 | -0.1034 | No |
| 49 | Tubb2a | 6757 | -0.065 | -0.1123 | No |
| 50 | Imp3 | 6803 | -0.069 | -0.1130 | No |
| 51 | Dnajc3 | 6872 | -0.074 | -0.1155 | No |
| 52 | Parn | 6907 | -0.076 | -0.1151 | No |
| 53 | Spcs1 | 7108 | -0.091 | -0.1276 | No |
| 54 | Pop4 | 7135 | -0.093 | -0.1258 | No |
| 55 | Eif4e | 7245 | -0.099 | -0.1306 | No |
| 56 | H2ax | 7419 | -0.111 | -0.1400 | No |
| 57 | Exosc5 | 7455 | -0.114 | -0.1382 | No |
| 58 | Atf4 | 7465 | -0.114 | -0.1342 | No |
| 59 | Nabp1 | 7520 | -0.118 | -0.1337 | No |
| 60 | Exosc4 | 7545 | -0.120 | -0.1307 | No |
| 61 | Gosr2 | 7562 | -0.121 | -0.1270 | No |
| 62 | Shc1 | 7575 | -0.121 | -0.1229 | No |
| 63 | Edem1 | 7634 | -0.125 | -0.1225 | No |
| 64 | Sec11a | 7931 | -0.148 | -0.1403 | No |
| 65 | Hspa9 | 7977 | -0.151 | -0.1377 | No |
| 66 | Calr | 7987 | -0.152 | -0.1322 | No |
| 67 | Fus | 8299 | -0.172 | -0.1502 | No |
| 68 | Atf6 | 8590 | -0.192 | -0.1658 | No |
| 69 | Nolc1 | 8776 | -0.207 | -0.1722 | No |
| 70 | Pdia5 | 8843 | -0.212 | -0.1688 | No |
| 71 | Eif2s1 | 8925 | -0.219 | -0.1663 | No |
| 72 | Nop56 | 9286 | -0.247 | -0.1853 | No |
| 73 | Srpr | 9388 | -0.256 | -0.1829 | No |
| 74 | Tatdn2 | 9533 | -0.267 | -0.1836 | No |
| 75 | Eef2 | 9932 | -0.302 | -0.2033 | Yes |
| 76 | Eif4ebp1 | 9951 | -0.304 | -0.1922 | Yes |
| 77 | Exosc9 | 9977 | -0.307 | -0.1816 | Yes |
| 78 | Xpot | 9987 | -0.308 | -0.1696 | Yes |
| 79 | Tspyl2 | 10039 | -0.313 | -0.1608 | Yes |
| 80 | Cebpg | 10227 | -0.332 | -0.1622 | Yes |
| 81 | Ssr1 | 10262 | -0.336 | -0.1511 | Yes |
| 82 | Banf1 | 10302 | -0.340 | -0.1402 | Yes |
| 83 | Rrp9 | 10406 | -0.351 | -0.1341 | Yes |
| 84 | Dcp1a | 10598 | -0.372 | -0.1342 | Yes |
| 85 | Herpud1 | 10756 | -0.389 | -0.1308 | Yes |
| 86 | Khsrp | 10798 | -0.395 | -0.1178 | Yes |
| 87 | Xbp1 | 10825 | -0.398 | -0.1035 | Yes |
| 88 | Exoc2 | 11058 | -0.427 | -0.1046 | Yes |
| 89 | Lsm4 | 11182 | -0.443 | -0.0963 | Yes |
| 90 | Exosc1 | 11209 | -0.448 | -0.0799 | Yes |
| 91 | Pdia6 | 11325 | -0.466 | -0.0700 | Yes |
| 92 | Mthfd2 | 11407 | -0.478 | -0.0568 | Yes |
| 93 | Mtrex | 11720 | -0.543 | -0.0596 | Yes |
| 94 | Cxxc1 | 11814 | -0.566 | -0.0438 | Yes |
| 95 | Nfya | 12117 | -0.677 | -0.0403 | Yes |
| 96 | Nfyb | 12269 | -0.788 | -0.0200 | Yes |
| 97 | Ern1 | 12299 | -0.819 | 0.0115 | Yes |