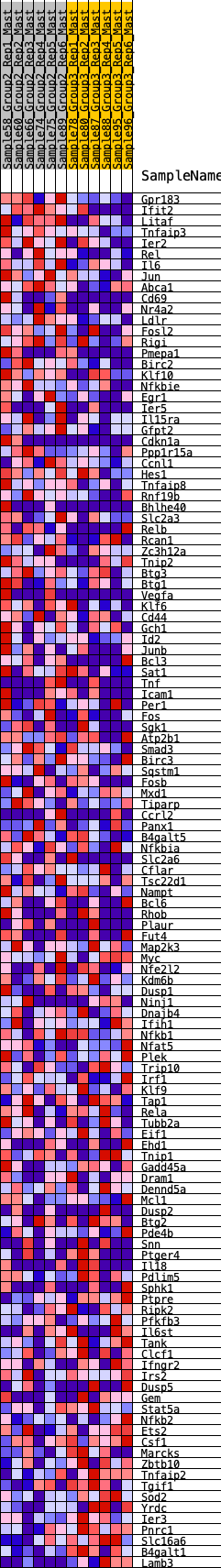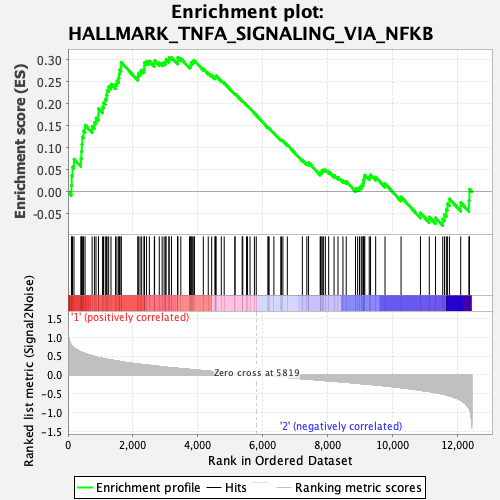
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.30553603 |
| Normalized Enrichment Score (NES) | 1.1781276 |
| Nominal p-value | 0.23856859 |
| FDR q-value | 0.9678935 |
| FWER p-Value | 0.941 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpr183 | 106 | 0.784 | 0.0144 | Yes |
| 2 | Ifit2 | 115 | 0.776 | 0.0366 | Yes |
| 3 | Litaf | 138 | 0.750 | 0.0569 | Yes |
| 4 | Tnfaip3 | 184 | 0.717 | 0.0743 | Yes |
| 5 | Ier2 | 396 | 0.611 | 0.0752 | Yes |
| 6 | Rel | 412 | 0.605 | 0.0918 | Yes |
| 7 | Il6 | 428 | 0.598 | 0.1081 | Yes |
| 8 | Jun | 441 | 0.595 | 0.1247 | Yes |
| 9 | Abca1 | 479 | 0.584 | 0.1388 | Yes |
| 10 | Cd69 | 526 | 0.566 | 0.1518 | Yes |
| 11 | Nr4a2 | 743 | 0.508 | 0.1492 | Yes |
| 12 | Ldlr | 814 | 0.489 | 0.1579 | Yes |
| 13 | Fosl2 | 863 | 0.478 | 0.1680 | Yes |
| 14 | Rigi | 938 | 0.462 | 0.1756 | Yes |
| 15 | Pmepa1 | 940 | 0.462 | 0.1891 | Yes |
| 16 | Birc2 | 1065 | 0.438 | 0.1920 | Yes |
| 17 | Klf10 | 1095 | 0.435 | 0.2024 | Yes |
| 18 | Nfkbie | 1149 | 0.425 | 0.2106 | Yes |
| 19 | Egr1 | 1183 | 0.421 | 0.2203 | Yes |
| 20 | Ier5 | 1204 | 0.417 | 0.2310 | Yes |
| 21 | Il15ra | 1253 | 0.408 | 0.2391 | Yes |
| 22 | Gfpt2 | 1325 | 0.397 | 0.2450 | Yes |
| 23 | Cdkn1a | 1465 | 0.376 | 0.2448 | Yes |
| 24 | Ppp1r15a | 1507 | 0.370 | 0.2523 | Yes |
| 25 | Ccnl1 | 1562 | 0.361 | 0.2586 | Yes |
| 26 | Hes1 | 1576 | 0.360 | 0.2681 | Yes |
| 27 | Tnfaip8 | 1590 | 0.359 | 0.2776 | Yes |
| 28 | Rnf19b | 1636 | 0.353 | 0.2843 | Yes |
| 29 | Bhlhe40 | 1638 | 0.352 | 0.2946 | Yes |
| 30 | Slc2a3 | 2150 | 0.288 | 0.2616 | Yes |
| 31 | Relb | 2165 | 0.286 | 0.2689 | Yes |
| 32 | Rcan1 | 2219 | 0.281 | 0.2729 | Yes |
| 33 | Zc3h12a | 2269 | 0.277 | 0.2770 | Yes |
| 34 | Tnip2 | 2342 | 0.271 | 0.2791 | Yes |
| 35 | Btg3 | 2353 | 0.270 | 0.2863 | Yes |
| 36 | Btg1 | 2355 | 0.269 | 0.2941 | Yes |
| 37 | Vegfa | 2420 | 0.264 | 0.2967 | Yes |
| 38 | Klf6 | 2507 | 0.257 | 0.2973 | Yes |
| 39 | Cd44 | 2659 | 0.239 | 0.2920 | Yes |
| 40 | Gch1 | 2668 | 0.239 | 0.2984 | Yes |
| 41 | Id2 | 2812 | 0.225 | 0.2934 | Yes |
| 42 | Junb | 2901 | 0.216 | 0.2926 | Yes |
| 43 | Bcl3 | 2961 | 0.210 | 0.2940 | Yes |
| 44 | Sat1 | 3013 | 0.206 | 0.2959 | Yes |
| 45 | Tnf | 3018 | 0.205 | 0.3017 | Yes |
| 46 | Icam1 | 3110 | 0.197 | 0.3001 | Yes |
| 47 | Per1 | 3115 | 0.197 | 0.3055 | Yes |
| 48 | Fos | 3188 | 0.190 | 0.3053 | No |
| 49 | Sgk1 | 3378 | 0.176 | 0.2951 | No |
| 50 | Atp2b1 | 3379 | 0.176 | 0.3003 | No |
| 51 | Smad3 | 3380 | 0.176 | 0.3055 | No |
| 52 | Birc3 | 3476 | 0.167 | 0.3027 | No |
| 53 | Sqstm1 | 3744 | 0.147 | 0.2853 | No |
| 54 | Fosb | 3782 | 0.144 | 0.2865 | No |
| 55 | Mxd1 | 3789 | 0.143 | 0.2903 | No |
| 56 | Tiparp | 3796 | 0.143 | 0.2940 | No |
| 57 | Ccrl2 | 3846 | 0.140 | 0.2941 | No |
| 58 | Panx1 | 3852 | 0.139 | 0.2978 | No |
| 59 | B4galt5 | 3896 | 0.135 | 0.2983 | No |
| 60 | Nfkbia | 4169 | 0.114 | 0.2795 | No |
| 61 | Slc2a6 | 4321 | 0.103 | 0.2703 | No |
| 62 | Cflar | 4426 | 0.093 | 0.2646 | No |
| 63 | Tsc22d1 | 4528 | 0.084 | 0.2589 | No |
| 64 | Nampt | 4541 | 0.084 | 0.2604 | No |
| 65 | Bcl6 | 4549 | 0.083 | 0.2622 | No |
| 66 | Rhob | 4562 | 0.081 | 0.2636 | No |
| 67 | Plaur | 4723 | 0.072 | 0.2528 | No |
| 68 | Fut4 | 4812 | 0.067 | 0.2476 | No |
| 69 | Map2k3 | 5142 | 0.044 | 0.2222 | No |
| 70 | Myc | 5145 | 0.044 | 0.2233 | No |
| 71 | Nfe2l2 | 5366 | 0.030 | 0.2063 | No |
| 72 | Kdm6b | 5385 | 0.028 | 0.2057 | No |
| 73 | Dusp1 | 5501 | 0.021 | 0.1970 | No |
| 74 | Ninj1 | 5531 | 0.018 | 0.1952 | No |
| 75 | Dnajb4 | 5608 | 0.014 | 0.1894 | No |
| 76 | Ifih1 | 5743 | 0.005 | 0.1786 | No |
| 77 | Nfkb1 | 5800 | 0.002 | 0.1742 | No |
| 78 | Nfat5 | 6160 | -0.023 | 0.1457 | No |
| 79 | Plek | 6172 | -0.024 | 0.1455 | No |
| 80 | Trip10 | 6196 | -0.026 | 0.1444 | No |
| 81 | Irf1 | 6343 | -0.035 | 0.1335 | No |
| 82 | Klf9 | 6555 | -0.051 | 0.1179 | No |
| 83 | Tap1 | 6581 | -0.053 | 0.1174 | No |
| 84 | Rela | 6621 | -0.055 | 0.1159 | No |
| 85 | Tubb2a | 6757 | -0.065 | 0.1068 | No |
| 86 | Eif1 | 7220 | -0.098 | 0.0722 | No |
| 87 | Ehd1 | 7357 | -0.107 | 0.0643 | No |
| 88 | Tnip1 | 7408 | -0.111 | 0.0635 | No |
| 89 | Gadd45a | 7411 | -0.111 | 0.0666 | No |
| 90 | Dram1 | 7769 | -0.135 | 0.0416 | No |
| 91 | Dennd5a | 7780 | -0.136 | 0.0448 | No |
| 92 | Mcl1 | 7824 | -0.140 | 0.0454 | No |
| 93 | Dusp2 | 7828 | -0.140 | 0.0493 | No |
| 94 | Btg2 | 7875 | -0.144 | 0.0498 | No |
| 95 | Pde4b | 7930 | -0.148 | 0.0497 | No |
| 96 | Snn | 8031 | -0.155 | 0.0462 | No |
| 97 | Ptger4 | 8197 | -0.165 | 0.0376 | No |
| 98 | Il18 | 8318 | -0.174 | 0.0330 | No |
| 99 | Pdlim5 | 8471 | -0.184 | 0.0261 | No |
| 100 | Sphk1 | 8572 | -0.191 | 0.0236 | No |
| 101 | Ptpre | 8858 | -0.213 | 0.0067 | No |
| 102 | Ripk2 | 8923 | -0.218 | 0.0079 | No |
| 103 | Pfkfb3 | 8981 | -0.224 | 0.0099 | No |
| 104 | Il6st | 9033 | -0.227 | 0.0124 | No |
| 105 | Tank | 9061 | -0.229 | 0.0170 | No |
| 106 | Clcf1 | 9098 | -0.232 | 0.0209 | No |
| 107 | Ifngr2 | 9105 | -0.233 | 0.0273 | No |
| 108 | Irs2 | 9124 | -0.234 | 0.0327 | No |
| 109 | Dusp5 | 9144 | -0.236 | 0.0381 | No |
| 110 | Gem | 9281 | -0.246 | 0.0343 | No |
| 111 | Stat5a | 9321 | -0.251 | 0.0385 | No |
| 112 | Nfkb2 | 9479 | -0.262 | 0.0335 | No |
| 113 | Ets2 | 9767 | -0.287 | 0.0186 | No |
| 114 | Csf1 | 10261 | -0.336 | -0.0116 | No |
| 115 | Marcks | 10859 | -0.403 | -0.0482 | No |
| 116 | Zbtb10 | 11131 | -0.436 | -0.0573 | No |
| 117 | Tnfaip2 | 11323 | -0.466 | -0.0591 | No |
| 118 | Tgif1 | 11545 | -0.502 | -0.0623 | No |
| 119 | Sod2 | 11599 | -0.513 | -0.0515 | No |
| 120 | Yrdc | 11656 | -0.527 | -0.0406 | No |
| 121 | Ier3 | 11689 | -0.534 | -0.0274 | No |
| 122 | Pnrc1 | 11751 | -0.550 | -0.0162 | No |
| 123 | Slc16a6 | 12099 | -0.669 | -0.0247 | No |
| 124 | B4galt1 | 12352 | -0.873 | -0.0195 | No |
| 125 | Lamb3 | 12368 | -0.904 | 0.0059 | No |