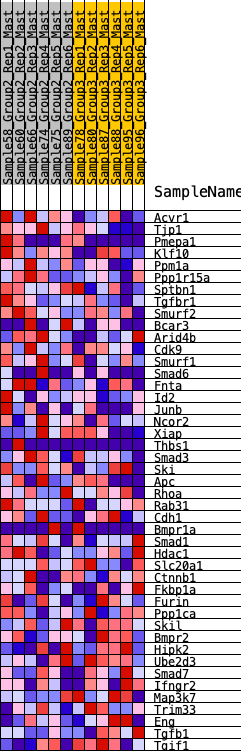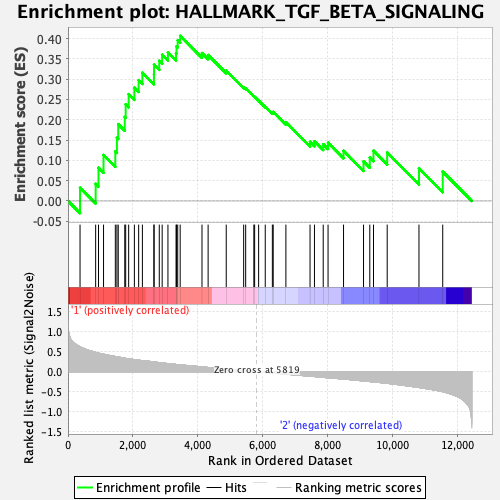
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.40649125 |
| Normalized Enrichment Score (NES) | 1.4442538 |
| Nominal p-value | 0.048728812 |
| FDR q-value | 0.7161267 |
| FWER p-Value | 0.471 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acvr1 | 371 | 0.619 | 0.0325 | Yes |
| 2 | Tjp1 | 852 | 0.480 | 0.0423 | Yes |
| 3 | Pmepa1 | 940 | 0.462 | 0.0818 | Yes |
| 4 | Klf10 | 1095 | 0.435 | 0.1133 | Yes |
| 5 | Ppm1a | 1458 | 0.378 | 0.1222 | Yes |
| 6 | Ppp1r15a | 1507 | 0.370 | 0.1557 | Yes |
| 7 | Sptbn1 | 1548 | 0.364 | 0.1892 | Yes |
| 8 | Tgfbr1 | 1749 | 0.335 | 0.2068 | Yes |
| 9 | Smurf2 | 1777 | 0.332 | 0.2381 | Yes |
| 10 | Bcar3 | 1869 | 0.320 | 0.2630 | Yes |
| 11 | Arid4b | 2045 | 0.299 | 0.2791 | Yes |
| 12 | Cdk9 | 2177 | 0.285 | 0.2973 | Yes |
| 13 | Smurf1 | 2292 | 0.275 | 0.3158 | Yes |
| 14 | Smad6 | 2648 | 0.241 | 0.3115 | Yes |
| 15 | Fnta | 2651 | 0.240 | 0.3356 | Yes |
| 16 | Id2 | 2812 | 0.225 | 0.3454 | Yes |
| 17 | Junb | 2901 | 0.216 | 0.3600 | Yes |
| 18 | Ncor2 | 3079 | 0.199 | 0.3658 | Yes |
| 19 | Xiap | 3329 | 0.179 | 0.3638 | Yes |
| 20 | Thbs1 | 3344 | 0.178 | 0.3806 | Yes |
| 21 | Smad3 | 3380 | 0.176 | 0.3955 | Yes |
| 22 | Ski | 3456 | 0.169 | 0.4065 | Yes |
| 23 | Apc | 4128 | 0.117 | 0.3641 | No |
| 24 | Rhoa | 4317 | 0.103 | 0.3594 | No |
| 25 | Rab31 | 4875 | 0.062 | 0.3207 | No |
| 26 | Cdh1 | 5412 | 0.027 | 0.2802 | No |
| 27 | Bmpr1a | 5474 | 0.022 | 0.2775 | No |
| 28 | Smad1 | 5726 | 0.006 | 0.2579 | No |
| 29 | Hdac1 | 5750 | 0.004 | 0.2565 | No |
| 30 | Slc20a1 | 5873 | -0.003 | 0.2470 | No |
| 31 | Ctnnb1 | 6077 | -0.017 | 0.2323 | No |
| 32 | Fkbp1a | 6296 | -0.032 | 0.2180 | No |
| 33 | Furin | 6322 | -0.034 | 0.2194 | No |
| 34 | Ppp1ca | 6713 | -0.062 | 0.1941 | No |
| 35 | Skil | 7457 | -0.114 | 0.1457 | No |
| 36 | Bmpr2 | 7593 | -0.122 | 0.1471 | No |
| 37 | Hipk2 | 7863 | -0.142 | 0.1398 | No |
| 38 | Ube2d3 | 8012 | -0.154 | 0.1434 | No |
| 39 | Smad7 | 8489 | -0.185 | 0.1236 | No |
| 40 | Ifngr2 | 9105 | -0.233 | 0.0975 | No |
| 41 | Map3k7 | 9298 | -0.248 | 0.1070 | No |
| 42 | Trim33 | 9412 | -0.258 | 0.1239 | No |
| 43 | Eng | 9834 | -0.292 | 0.1194 | No |
| 44 | Tgfb1 | 10813 | -0.397 | 0.0806 | No |
| 45 | Tgif1 | 11545 | -0.502 | 0.0723 | No |