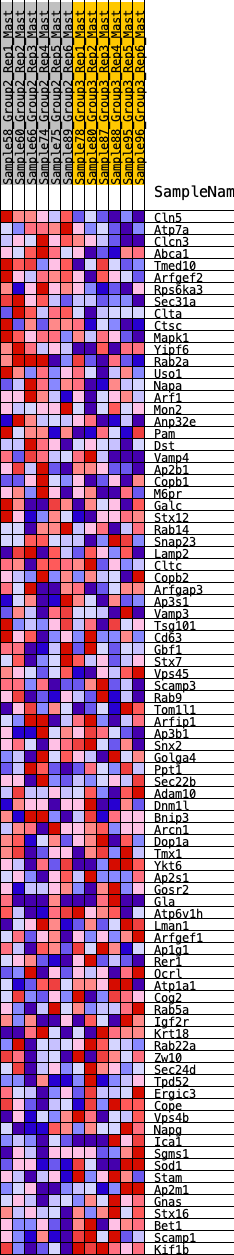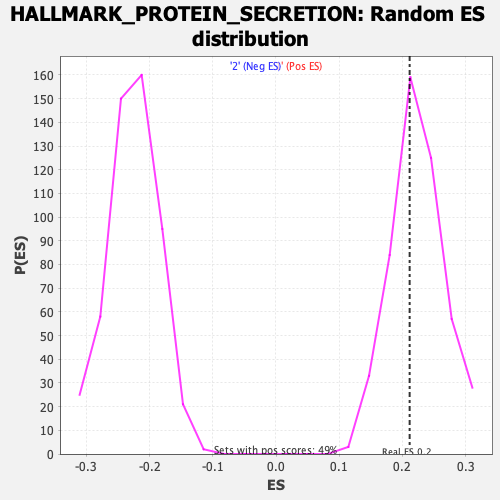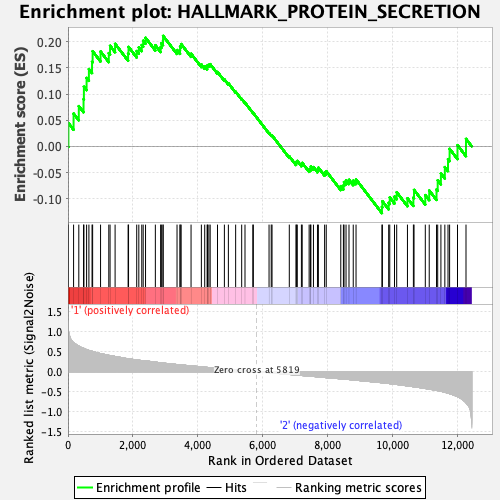
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.21138528 |
| Normalized Enrichment Score (NES) | 0.9499986 |
| Nominal p-value | 0.599182 |
| FDR q-value | 0.91145504 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cln5 | 13 | 1.072 | 0.0447 | Yes |
| 2 | Atp7a | 172 | 0.724 | 0.0628 | Yes |
| 3 | Clcn3 | 333 | 0.638 | 0.0771 | Yes |
| 4 | Abca1 | 479 | 0.584 | 0.0903 | Yes |
| 5 | Tmed10 | 490 | 0.581 | 0.1142 | Yes |
| 6 | Arfgef2 | 573 | 0.549 | 0.1310 | Yes |
| 7 | Rps6ka3 | 644 | 0.530 | 0.1480 | Yes |
| 8 | Sec31a | 741 | 0.508 | 0.1619 | Yes |
| 9 | Clta | 759 | 0.504 | 0.1820 | Yes |
| 10 | Ctsc | 1003 | 0.450 | 0.1816 | Yes |
| 11 | Mapk1 | 1257 | 0.407 | 0.1784 | Yes |
| 12 | Yipf6 | 1296 | 0.402 | 0.1925 | Yes |
| 13 | Rab2a | 1451 | 0.379 | 0.1962 | Yes |
| 14 | Uso1 | 1852 | 0.322 | 0.1776 | Yes |
| 15 | Napa | 1865 | 0.320 | 0.1903 | Yes |
| 16 | Arf1 | 2117 | 0.293 | 0.1824 | Yes |
| 17 | Mon2 | 2182 | 0.285 | 0.1894 | Yes |
| 18 | Anp32e | 2273 | 0.276 | 0.1939 | Yes |
| 19 | Pam | 2316 | 0.273 | 0.2022 | Yes |
| 20 | Dst | 2389 | 0.267 | 0.2077 | Yes |
| 21 | Vamp4 | 2691 | 0.238 | 0.1935 | Yes |
| 22 | Ap2b1 | 2854 | 0.221 | 0.1898 | Yes |
| 23 | Copb1 | 2876 | 0.218 | 0.1974 | Yes |
| 24 | M6pr | 2927 | 0.213 | 0.2025 | Yes |
| 25 | Galc | 2930 | 0.213 | 0.2114 | Yes |
| 26 | Stx12 | 3359 | 0.178 | 0.1843 | No |
| 27 | Rab14 | 3452 | 0.169 | 0.1841 | No |
| 28 | Snap23 | 3454 | 0.169 | 0.1912 | No |
| 29 | Lamp2 | 3486 | 0.167 | 0.1958 | No |
| 30 | Cltc | 3791 | 0.143 | 0.1773 | No |
| 31 | Copb2 | 4109 | 0.118 | 0.1567 | No |
| 32 | Arfgap3 | 4212 | 0.112 | 0.1532 | No |
| 33 | Ap3s1 | 4286 | 0.105 | 0.1518 | No |
| 34 | Vamp3 | 4298 | 0.104 | 0.1553 | No |
| 35 | Tsg101 | 4340 | 0.101 | 0.1563 | No |
| 36 | Cd63 | 4380 | 0.097 | 0.1573 | No |
| 37 | Gbf1 | 4606 | 0.078 | 0.1424 | No |
| 38 | Stx7 | 4818 | 0.066 | 0.1282 | No |
| 39 | Vps45 | 4941 | 0.059 | 0.1208 | No |
| 40 | Scamp3 | 5165 | 0.043 | 0.1046 | No |
| 41 | Rab9 | 5351 | 0.030 | 0.0909 | No |
| 42 | Tom1l1 | 5453 | 0.023 | 0.0837 | No |
| 43 | Arfip1 | 5694 | 0.008 | 0.0646 | No |
| 44 | Ap3b1 | 5710 | 0.007 | 0.0637 | No |
| 45 | Snx2 | 6194 | -0.025 | 0.0257 | No |
| 46 | Golga4 | 6261 | -0.030 | 0.0216 | No |
| 47 | Ppt1 | 6286 | -0.032 | 0.0210 | No |
| 48 | Sec22b | 6818 | -0.070 | -0.0190 | No |
| 49 | Adam10 | 7024 | -0.085 | -0.0320 | No |
| 50 | Dnm1l | 7037 | -0.085 | -0.0293 | No |
| 51 | Bnip3 | 7064 | -0.088 | -0.0276 | No |
| 52 | Arcn1 | 7191 | -0.096 | -0.0337 | No |
| 53 | Dop1a | 7215 | -0.097 | -0.0314 | No |
| 54 | Tmx1 | 7429 | -0.112 | -0.0439 | No |
| 55 | Ykt6 | 7468 | -0.115 | -0.0421 | No |
| 56 | Ap2s1 | 7483 | -0.115 | -0.0383 | No |
| 57 | Gosr2 | 7562 | -0.121 | -0.0395 | No |
| 58 | Gla | 7686 | -0.129 | -0.0439 | No |
| 59 | Atp6v1h | 7714 | -0.131 | -0.0405 | No |
| 60 | Lman1 | 7906 | -0.146 | -0.0497 | No |
| 61 | Arfgef1 | 7956 | -0.150 | -0.0473 | No |
| 62 | Ap1g1 | 8404 | -0.179 | -0.0758 | No |
| 63 | Rer1 | 8485 | -0.185 | -0.0744 | No |
| 64 | Ocrl | 8504 | -0.186 | -0.0680 | No |
| 65 | Atp1a1 | 8566 | -0.191 | -0.0648 | No |
| 66 | Cog2 | 8657 | -0.198 | -0.0636 | No |
| 67 | Rab5a | 8788 | -0.209 | -0.0652 | No |
| 68 | Igf2r | 8874 | -0.214 | -0.0630 | No |
| 69 | Krt18 | 9672 | -0.278 | -0.1156 | No |
| 70 | Rab22a | 9686 | -0.280 | -0.1048 | No |
| 71 | Zw10 | 9880 | -0.297 | -0.1077 | No |
| 72 | Sec24d | 9913 | -0.300 | -0.0975 | No |
| 73 | Tpd52 | 10059 | -0.315 | -0.0958 | No |
| 74 | Ergic3 | 10127 | -0.321 | -0.0876 | No |
| 75 | Cope | 10458 | -0.356 | -0.0991 | No |
| 76 | Vps4b | 10641 | -0.377 | -0.0977 | No |
| 77 | Napg | 10661 | -0.379 | -0.0831 | No |
| 78 | Ica1 | 11008 | -0.422 | -0.0931 | No |
| 79 | Sgms1 | 11129 | -0.436 | -0.0842 | No |
| 80 | Sod1 | 11354 | -0.471 | -0.0822 | No |
| 81 | Stam | 11390 | -0.475 | -0.0648 | No |
| 82 | Ap2m1 | 11489 | -0.492 | -0.0517 | No |
| 83 | Gnas | 11610 | -0.516 | -0.0394 | No |
| 84 | Stx16 | 11708 | -0.540 | -0.0243 | No |
| 85 | Bet1 | 11757 | -0.551 | -0.0046 | No |
| 86 | Scamp1 | 12000 | -0.624 | 0.0024 | No |
| 87 | Kif1b | 12262 | -0.779 | 0.0145 | No |