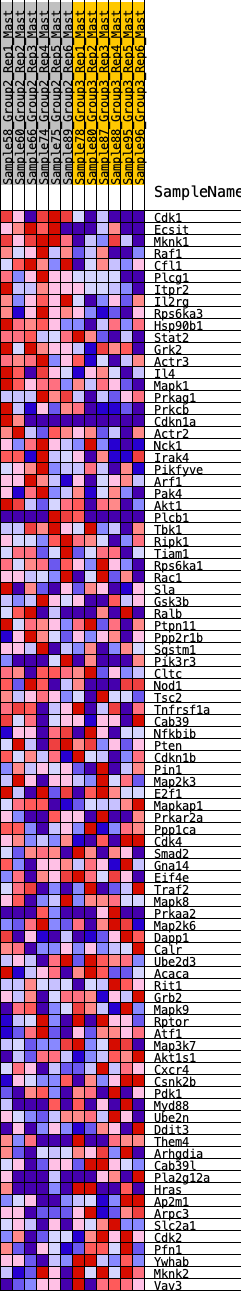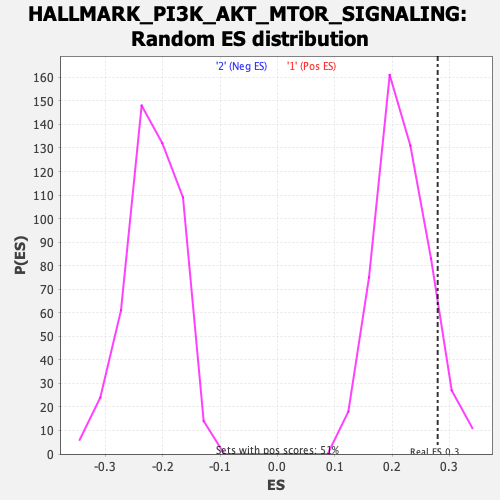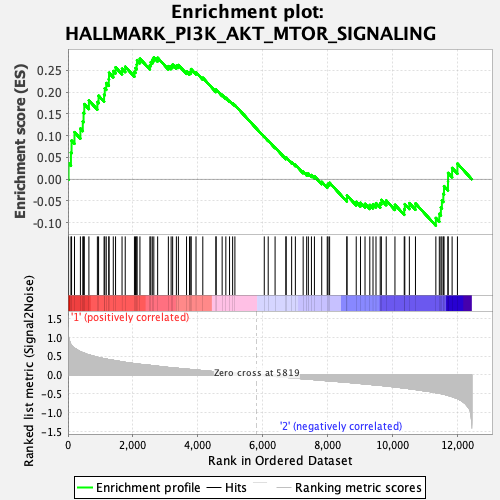
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.27948114 |
| Normalized Enrichment Score (NES) | 1.2734929 |
| Nominal p-value | 0.09486166 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.833 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdk1 | 18 | 1.017 | 0.0366 | Yes |
| 2 | Ecsit | 91 | 0.804 | 0.0608 | Yes |
| 3 | Mknk1 | 110 | 0.783 | 0.0886 | Yes |
| 4 | Raf1 | 200 | 0.708 | 0.1079 | Yes |
| 5 | Cfl1 | 382 | 0.616 | 0.1163 | Yes |
| 6 | Plcg1 | 456 | 0.590 | 0.1324 | Yes |
| 7 | Itpr2 | 480 | 0.584 | 0.1524 | Yes |
| 8 | Il2rg | 502 | 0.578 | 0.1723 | Yes |
| 9 | Rps6ka3 | 644 | 0.530 | 0.1807 | Yes |
| 10 | Hsp90b1 | 905 | 0.469 | 0.1772 | Yes |
| 11 | Stat2 | 942 | 0.462 | 0.1916 | Yes |
| 12 | Grk2 | 1112 | 0.432 | 0.1940 | Yes |
| 13 | Actr3 | 1133 | 0.429 | 0.2085 | Yes |
| 14 | Il4 | 1178 | 0.421 | 0.2207 | Yes |
| 15 | Mapk1 | 1257 | 0.407 | 0.2296 | Yes |
| 16 | Prkag1 | 1264 | 0.406 | 0.2443 | Yes |
| 17 | Prkcb | 1395 | 0.387 | 0.2482 | Yes |
| 18 | Cdkn1a | 1465 | 0.376 | 0.2567 | Yes |
| 19 | Actr2 | 1665 | 0.348 | 0.2536 | Yes |
| 20 | Nck1 | 1763 | 0.333 | 0.2582 | Yes |
| 21 | Irak4 | 2047 | 0.299 | 0.2465 | Yes |
| 22 | Pikfyve | 2079 | 0.296 | 0.2550 | Yes |
| 23 | Arf1 | 2117 | 0.293 | 0.2630 | Yes |
| 24 | Pak4 | 2127 | 0.292 | 0.2732 | Yes |
| 25 | Akt1 | 2216 | 0.281 | 0.2766 | Yes |
| 26 | Plcb1 | 2523 | 0.256 | 0.2614 | Yes |
| 27 | Tbk1 | 2551 | 0.251 | 0.2686 | Yes |
| 28 | Ripk1 | 2597 | 0.247 | 0.2742 | Yes |
| 29 | Tiam1 | 2644 | 0.242 | 0.2795 | Yes |
| 30 | Rps6ka1 | 2762 | 0.230 | 0.2786 | No |
| 31 | Rac1 | 3092 | 0.198 | 0.2594 | No |
| 32 | Sla | 3177 | 0.191 | 0.2597 | No |
| 33 | Gsk3b | 3224 | 0.187 | 0.2630 | No |
| 34 | Ralb | 3339 | 0.178 | 0.2604 | No |
| 35 | Ptpn11 | 3401 | 0.174 | 0.2620 | No |
| 36 | Ppp2r1b | 3652 | 0.154 | 0.2475 | No |
| 37 | Sqstm1 | 3744 | 0.147 | 0.2456 | No |
| 38 | Pik3r3 | 3787 | 0.143 | 0.2476 | No |
| 39 | Cltc | 3791 | 0.143 | 0.2527 | No |
| 40 | Nod1 | 3944 | 0.131 | 0.2453 | No |
| 41 | Tsc2 | 4153 | 0.115 | 0.2328 | No |
| 42 | Tnfrsf1a | 4559 | 0.082 | 0.2030 | No |
| 43 | Cab39 | 4561 | 0.081 | 0.2060 | No |
| 44 | Nfkbib | 4749 | 0.070 | 0.1935 | No |
| 45 | Pten | 4863 | 0.063 | 0.1867 | No |
| 46 | Cdkn1b | 4977 | 0.056 | 0.1796 | No |
| 47 | Pin1 | 5072 | 0.050 | 0.1739 | No |
| 48 | Map2k3 | 5142 | 0.044 | 0.1699 | No |
| 49 | E2f1 | 6047 | -0.015 | 0.0973 | No |
| 50 | Mapkap1 | 6168 | -0.023 | 0.0885 | No |
| 51 | Prkar2a | 6380 | -0.038 | 0.0728 | No |
| 52 | Ppp1ca | 6713 | -0.062 | 0.0483 | No |
| 53 | Cdk4 | 6720 | -0.062 | 0.0501 | No |
| 54 | Smad2 | 6890 | -0.075 | 0.0392 | No |
| 55 | Gna14 | 7005 | -0.083 | 0.0331 | No |
| 56 | Eif4e | 7245 | -0.099 | 0.0175 | No |
| 57 | Traf2 | 7353 | -0.107 | 0.0128 | No |
| 58 | Mapk8 | 7405 | -0.110 | 0.0128 | No |
| 59 | Prkaa2 | 7500 | -0.116 | 0.0096 | No |
| 60 | Map2k6 | 7592 | -0.122 | 0.0068 | No |
| 61 | Dapp1 | 7817 | -0.139 | -0.0062 | No |
| 62 | Calr | 7987 | -0.152 | -0.0142 | No |
| 63 | Ube2d3 | 8012 | -0.154 | -0.0104 | No |
| 64 | Acaca | 8060 | -0.157 | -0.0083 | No |
| 65 | Rit1 | 8591 | -0.192 | -0.0440 | No |
| 66 | Grb2 | 8595 | -0.192 | -0.0371 | No |
| 67 | Mapk9 | 8880 | -0.215 | -0.0520 | No |
| 68 | Rptor | 9012 | -0.226 | -0.0542 | No |
| 69 | Atf1 | 9151 | -0.236 | -0.0565 | No |
| 70 | Map3k7 | 9298 | -0.248 | -0.0591 | No |
| 71 | Akt1s1 | 9397 | -0.257 | -0.0574 | No |
| 72 | Cxcr4 | 9489 | -0.263 | -0.0549 | No |
| 73 | Csnk2b | 9619 | -0.274 | -0.0552 | No |
| 74 | Pdk1 | 9654 | -0.276 | -0.0476 | No |
| 75 | Myd88 | 9804 | -0.289 | -0.0488 | No |
| 76 | Ube2n | 10073 | -0.316 | -0.0587 | No |
| 77 | Ddit3 | 10359 | -0.346 | -0.0688 | No |
| 78 | Them4 | 10377 | -0.347 | -0.0572 | No |
| 79 | Arhgdia | 10517 | -0.363 | -0.0549 | No |
| 80 | Cab39l | 10705 | -0.384 | -0.0557 | No |
| 81 | Pla2g12a | 11333 | -0.467 | -0.0890 | No |
| 82 | Hras | 11440 | -0.483 | -0.0795 | No |
| 83 | Ap2m1 | 11489 | -0.492 | -0.0650 | No |
| 84 | Arpc3 | 11520 | -0.497 | -0.0488 | No |
| 85 | Slc2a1 | 11564 | -0.504 | -0.0334 | No |
| 86 | Cdk2 | 11585 | -0.509 | -0.0160 | No |
| 87 | Pfn1 | 11707 | -0.539 | -0.0057 | No |
| 88 | Ywhab | 11710 | -0.540 | 0.0144 | No |
| 89 | Mknk2 | 11834 | -0.572 | 0.0258 | No |
| 90 | Vav3 | 11998 | -0.623 | 0.0359 | No |