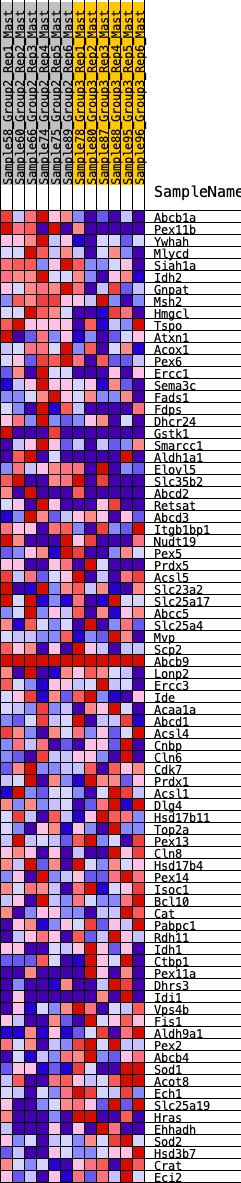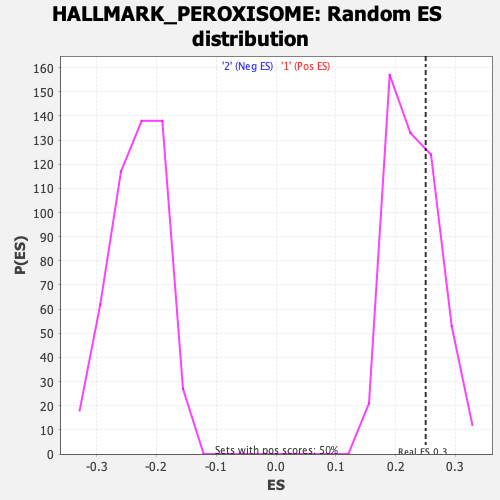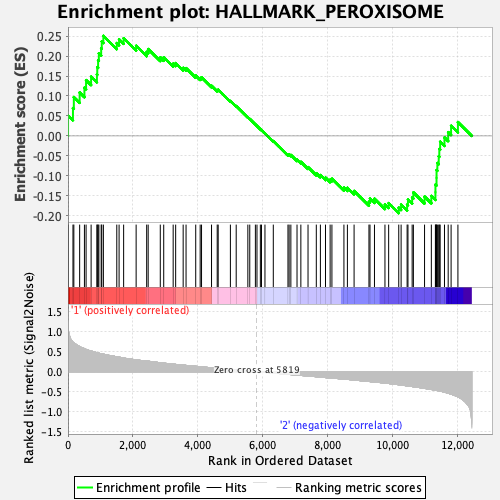
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.2504686 |
| Normalized Enrichment Score (NES) | 1.0913117 |
| Nominal p-value | 0.31 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcb1a | 2 | 1.245 | 0.0513 | Yes |
| 2 | Pex11b | 153 | 0.735 | 0.0696 | Yes |
| 3 | Ywhah | 180 | 0.718 | 0.0972 | Yes |
| 4 | Mlycd | 359 | 0.626 | 0.1087 | Yes |
| 5 | Siah1a | 506 | 0.576 | 0.1207 | Yes |
| 6 | Idh2 | 557 | 0.556 | 0.1397 | Yes |
| 7 | Gnpat | 712 | 0.515 | 0.1485 | Yes |
| 8 | Msh2 | 891 | 0.472 | 0.1536 | Yes |
| 9 | Hmgcl | 902 | 0.469 | 0.1722 | Yes |
| 10 | Tspo | 928 | 0.465 | 0.1894 | Yes |
| 11 | Atxn1 | 951 | 0.460 | 0.2067 | Yes |
| 12 | Acox1 | 1022 | 0.446 | 0.2194 | Yes |
| 13 | Pex6 | 1040 | 0.444 | 0.2364 | Yes |
| 14 | Ercc1 | 1090 | 0.435 | 0.2505 | Yes |
| 15 | Sema3c | 1503 | 0.371 | 0.2325 | No |
| 16 | Fads1 | 1574 | 0.360 | 0.2417 | No |
| 17 | Fdps | 1714 | 0.340 | 0.2445 | No |
| 18 | Dhcr24 | 2099 | 0.294 | 0.2256 | No |
| 19 | Gstk1 | 2424 | 0.264 | 0.2103 | No |
| 20 | Smarcc1 | 2470 | 0.260 | 0.2175 | No |
| 21 | Aldh1a1 | 2843 | 0.222 | 0.1965 | No |
| 22 | Elovl5 | 2949 | 0.211 | 0.1968 | No |
| 23 | Slc35b2 | 3240 | 0.186 | 0.1810 | No |
| 24 | Abcd2 | 3317 | 0.180 | 0.1823 | No |
| 25 | Retsat | 3548 | 0.161 | 0.1704 | No |
| 26 | Abcd3 | 3637 | 0.154 | 0.1696 | No |
| 27 | Itgb1bp1 | 3935 | 0.132 | 0.1511 | No |
| 28 | Nudt19 | 4072 | 0.121 | 0.1450 | No |
| 29 | Pex5 | 4114 | 0.118 | 0.1466 | No |
| 30 | Prdx5 | 4422 | 0.093 | 0.1256 | No |
| 31 | Acsl5 | 4597 | 0.079 | 0.1148 | No |
| 32 | Slc23a2 | 4627 | 0.077 | 0.1157 | No |
| 33 | Slc25a17 | 5003 | 0.054 | 0.0875 | No |
| 34 | Abcc5 | 5182 | 0.042 | 0.0749 | No |
| 35 | Slc25a4 | 5541 | 0.018 | 0.0466 | No |
| 36 | Mvp | 5600 | 0.014 | 0.0426 | No |
| 37 | Scp2 | 5773 | 0.003 | 0.0287 | No |
| 38 | Abcb9 | 5822 | 0.000 | 0.0249 | No |
| 39 | Lonp2 | 5926 | -0.006 | 0.0168 | No |
| 40 | Ercc3 | 5961 | -0.009 | 0.0144 | No |
| 41 | Ide | 6068 | -0.017 | 0.0065 | No |
| 42 | Acaa1a | 6325 | -0.034 | -0.0128 | No |
| 43 | Abcd1 | 6777 | -0.067 | -0.0465 | No |
| 44 | Acsl4 | 6812 | -0.069 | -0.0464 | No |
| 45 | Cnbp | 6866 | -0.073 | -0.0476 | No |
| 46 | Cln6 | 7056 | -0.087 | -0.0593 | No |
| 47 | Cdk7 | 7174 | -0.095 | -0.0649 | No |
| 48 | Prdx1 | 7395 | -0.110 | -0.0781 | No |
| 49 | Acsl1 | 7650 | -0.126 | -0.0935 | No |
| 50 | Dlg4 | 7776 | -0.136 | -0.0980 | No |
| 51 | Hsd17b11 | 7933 | -0.148 | -0.1044 | No |
| 52 | Top2a | 8076 | -0.158 | -0.1094 | No |
| 53 | Pex13 | 8131 | -0.161 | -0.1071 | No |
| 54 | Cln8 | 8501 | -0.185 | -0.1293 | No |
| 55 | Hsd17b4 | 8608 | -0.193 | -0.1299 | No |
| 56 | Pex14 | 8814 | -0.211 | -0.1377 | No |
| 57 | Isoc1 | 9272 | -0.246 | -0.1645 | No |
| 58 | Bcl10 | 9304 | -0.249 | -0.1568 | No |
| 59 | Cat | 9442 | -0.260 | -0.1571 | No |
| 60 | Pabpc1 | 9765 | -0.287 | -0.1713 | No |
| 61 | Rdh11 | 9879 | -0.296 | -0.1682 | No |
| 62 | Idh1 | 10190 | -0.328 | -0.1797 | No |
| 63 | Ctbp1 | 10263 | -0.336 | -0.1716 | No |
| 64 | Pex11a | 10448 | -0.354 | -0.1719 | No |
| 65 | Dhrs3 | 10474 | -0.358 | -0.1591 | No |
| 66 | Idi1 | 10603 | -0.373 | -0.1540 | No |
| 67 | Vps4b | 10641 | -0.377 | -0.1414 | No |
| 68 | Fis1 | 10985 | -0.419 | -0.1518 | No |
| 69 | Aldh9a1 | 11194 | -0.446 | -0.1502 | No |
| 70 | Pex2 | 11320 | -0.466 | -0.1411 | No |
| 71 | Abcb4 | 11321 | -0.466 | -0.1218 | No |
| 72 | Sod1 | 11354 | -0.471 | -0.1049 | No |
| 73 | Acot8 | 11355 | -0.471 | -0.0854 | No |
| 74 | Ech1 | 11380 | -0.474 | -0.0678 | No |
| 75 | Slc25a19 | 11422 | -0.481 | -0.0512 | No |
| 76 | Hras | 11440 | -0.483 | -0.0326 | No |
| 77 | Ehhadh | 11466 | -0.487 | -0.0145 | No |
| 78 | Sod2 | 11599 | -0.513 | -0.0039 | No |
| 79 | Hsd3b7 | 11713 | -0.541 | 0.0093 | No |
| 80 | Crat | 11803 | -0.563 | 0.0254 | No |
| 81 | Eci2 | 12013 | -0.631 | 0.0346 | No |