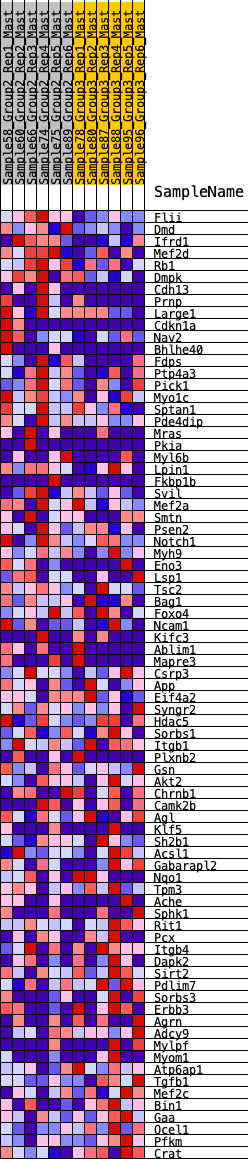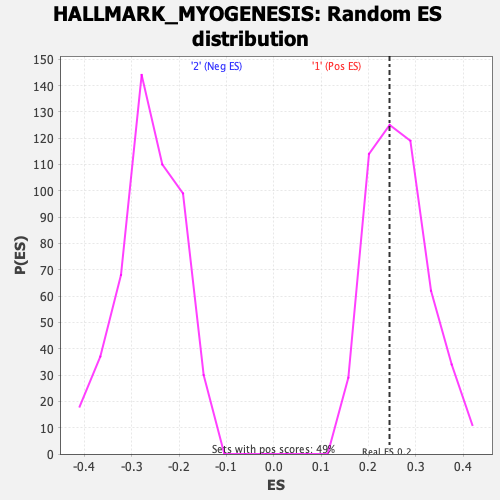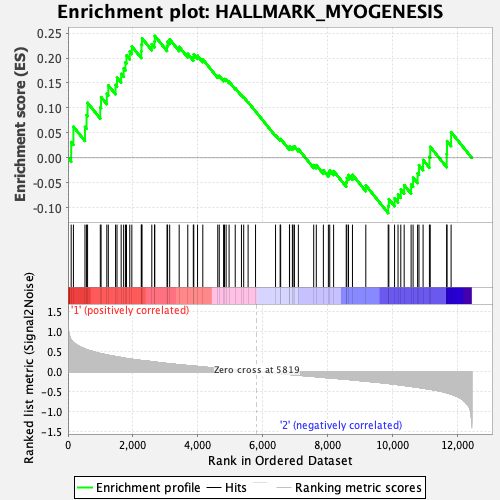
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.2444422 |
| Normalized Enrichment Score (NES) | 0.92885697 |
| Nominal p-value | 0.6052632 |
| FDR q-value | 0.8694317 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Flii | 98 | 0.793 | 0.0317 | Yes |
| 2 | Dmd | 167 | 0.727 | 0.0625 | Yes |
| 3 | Ifrd1 | 523 | 0.567 | 0.0621 | Yes |
| 4 | Mef2d | 572 | 0.549 | 0.0857 | Yes |
| 5 | Rb1 | 603 | 0.541 | 0.1103 | Yes |
| 6 | Dmpk | 995 | 0.452 | 0.1013 | Yes |
| 7 | Cdh13 | 1019 | 0.448 | 0.1218 | Yes |
| 8 | Prnp | 1196 | 0.419 | 0.1285 | Yes |
| 9 | Large1 | 1242 | 0.409 | 0.1453 | Yes |
| 10 | Cdkn1a | 1465 | 0.376 | 0.1462 | Yes |
| 11 | Nav2 | 1510 | 0.369 | 0.1611 | Yes |
| 12 | Bhlhe40 | 1638 | 0.352 | 0.1684 | Yes |
| 13 | Fdps | 1714 | 0.340 | 0.1793 | Yes |
| 14 | Ptp4a3 | 1770 | 0.332 | 0.1915 | Yes |
| 15 | Pick1 | 1802 | 0.327 | 0.2053 | Yes |
| 16 | Myo1c | 1903 | 0.317 | 0.2130 | Yes |
| 17 | Sptan1 | 1967 | 0.309 | 0.2234 | Yes |
| 18 | Pde4dip | 2258 | 0.278 | 0.2138 | Yes |
| 19 | Mras | 2263 | 0.277 | 0.2273 | Yes |
| 20 | Pkia | 2281 | 0.276 | 0.2397 | Yes |
| 21 | Myl6b | 2582 | 0.248 | 0.2279 | Yes |
| 22 | Lpin1 | 2665 | 0.239 | 0.2332 | Yes |
| 23 | Fkbp1b | 2674 | 0.238 | 0.2444 | Yes |
| 24 | Svil | 3051 | 0.202 | 0.2241 | No |
| 25 | Mef2a | 3068 | 0.200 | 0.2328 | No |
| 26 | Smtn | 3133 | 0.195 | 0.2374 | No |
| 27 | Psen2 | 3425 | 0.172 | 0.2225 | No |
| 28 | Notch1 | 3690 | 0.150 | 0.2086 | No |
| 29 | Myh9 | 3861 | 0.138 | 0.2018 | No |
| 30 | Eno3 | 3875 | 0.137 | 0.2076 | No |
| 31 | Lsp1 | 3991 | 0.129 | 0.2047 | No |
| 32 | Tsc2 | 4153 | 0.115 | 0.1975 | No |
| 33 | Bag1 | 4614 | 0.078 | 0.1641 | No |
| 34 | Foxo4 | 4660 | 0.075 | 0.1643 | No |
| 35 | Ncam1 | 4793 | 0.068 | 0.1570 | No |
| 36 | Kifc3 | 4822 | 0.066 | 0.1580 | No |
| 37 | Ablim1 | 4872 | 0.063 | 0.1572 | No |
| 38 | Mapre3 | 4963 | 0.057 | 0.1527 | No |
| 39 | Csrp3 | 5152 | 0.043 | 0.1397 | No |
| 40 | App | 5344 | 0.031 | 0.1258 | No |
| 41 | Eif4a2 | 5413 | 0.027 | 0.1216 | No |
| 42 | Syngr2 | 5549 | 0.018 | 0.1116 | No |
| 43 | Hdac5 | 5778 | 0.002 | 0.0932 | No |
| 44 | Sorbs1 | 6394 | -0.040 | 0.0455 | No |
| 45 | Itgb1 | 6536 | -0.050 | 0.0366 | No |
| 46 | Plxnb2 | 6552 | -0.051 | 0.0379 | No |
| 47 | Gsn | 6823 | -0.071 | 0.0196 | No |
| 48 | Akt2 | 6826 | -0.071 | 0.0230 | No |
| 49 | Chrnb1 | 6913 | -0.077 | 0.0199 | No |
| 50 | Camk2b | 6933 | -0.078 | 0.0222 | No |
| 51 | Agl | 6976 | -0.081 | 0.0229 | No |
| 52 | Klf5 | 7097 | -0.090 | 0.0177 | No |
| 53 | Sh2b1 | 7570 | -0.121 | -0.0144 | No |
| 54 | Acsl1 | 7650 | -0.126 | -0.0145 | No |
| 55 | Gabarapl2 | 7868 | -0.143 | -0.0249 | No |
| 56 | Nqo1 | 8022 | -0.154 | -0.0296 | No |
| 57 | Tpm3 | 8064 | -0.157 | -0.0251 | No |
| 58 | Ache | 8183 | -0.164 | -0.0264 | No |
| 59 | Sphk1 | 8572 | -0.191 | -0.0482 | No |
| 60 | Rit1 | 8591 | -0.192 | -0.0401 | No |
| 61 | Pcx | 8642 | -0.196 | -0.0343 | No |
| 62 | Itgb4 | 8764 | -0.206 | -0.0338 | No |
| 63 | Dapk2 | 9175 | -0.239 | -0.0550 | No |
| 64 | Sirt2 | 9865 | -0.295 | -0.0960 | No |
| 65 | Pdlim7 | 9885 | -0.297 | -0.0827 | No |
| 66 | Sorbs3 | 10061 | -0.315 | -0.0811 | No |
| 67 | Erbb3 | 10168 | -0.326 | -0.0734 | No |
| 68 | Agrn | 10253 | -0.335 | -0.0635 | No |
| 69 | Adcy9 | 10357 | -0.345 | -0.0546 | No |
| 70 | Mylpf | 10571 | -0.369 | -0.0534 | No |
| 71 | Myom1 | 10632 | -0.376 | -0.0394 | No |
| 72 | Atp6ap1 | 10771 | -0.391 | -0.0311 | No |
| 73 | Tgfb1 | 10813 | -0.397 | -0.0145 | No |
| 74 | Mef2c | 10940 | -0.414 | -0.0040 | No |
| 75 | Bin1 | 11135 | -0.437 | 0.0021 | No |
| 76 | Gaa | 11160 | -0.440 | 0.0222 | No |
| 77 | Ocel1 | 11667 | -0.529 | 0.0077 | No |
| 78 | Pfkm | 11677 | -0.533 | 0.0336 | No |
| 79 | Crat | 11803 | -0.563 | 0.0516 | No |