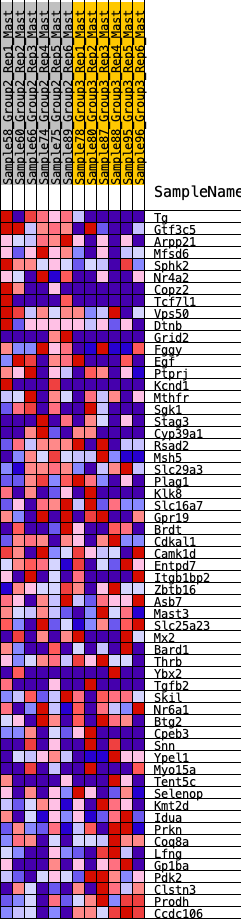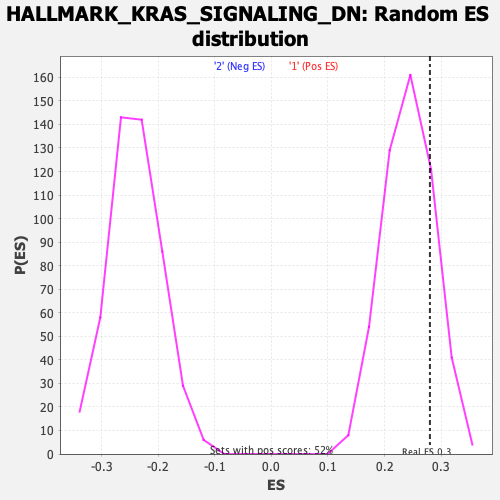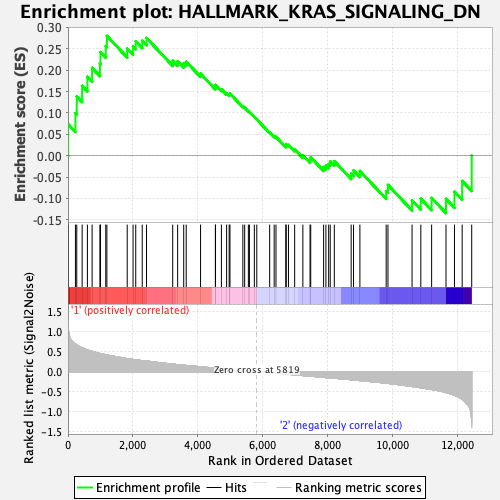
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.28021953 |
| Normalized Enrichment Score (NES) | 1.1519487 |
| Nominal p-value | 0.21235521 |
| FDR q-value | 0.9311552 |
| FWER p-Value | 0.963 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tg | 5 | 1.161 | 0.0735 | Yes |
| 2 | Gtf3c5 | 229 | 0.691 | 0.0995 | Yes |
| 3 | Arpp21 | 270 | 0.665 | 0.1387 | Yes |
| 4 | Mfsd6 | 436 | 0.596 | 0.1633 | Yes |
| 5 | Sphk2 | 598 | 0.542 | 0.1848 | Yes |
| 6 | Nr4a2 | 743 | 0.508 | 0.2055 | Yes |
| 7 | Copz2 | 983 | 0.453 | 0.2151 | Yes |
| 8 | Tcf7l1 | 1001 | 0.451 | 0.2424 | Yes |
| 9 | Vps50 | 1163 | 0.423 | 0.2563 | Yes |
| 10 | Dtnb | 1198 | 0.419 | 0.2802 | Yes |
| 11 | Grid2 | 1824 | 0.325 | 0.2505 | No |
| 12 | Fggy | 2004 | 0.305 | 0.2554 | No |
| 13 | Egf | 2087 | 0.295 | 0.2676 | No |
| 14 | Ptprj | 2287 | 0.275 | 0.2691 | No |
| 15 | Kcnd1 | 2419 | 0.264 | 0.2753 | No |
| 16 | Mthfr | 3226 | 0.187 | 0.2221 | No |
| 17 | Sgk1 | 3378 | 0.176 | 0.2211 | No |
| 18 | Stag3 | 3565 | 0.160 | 0.2163 | No |
| 19 | Cyp39a1 | 3642 | 0.154 | 0.2199 | No |
| 20 | Rsad2 | 4084 | 0.120 | 0.1919 | No |
| 21 | Msh5 | 4540 | 0.084 | 0.1605 | No |
| 22 | Slc29a3 | 4545 | 0.083 | 0.1655 | No |
| 23 | Plag1 | 4726 | 0.072 | 0.1555 | No |
| 24 | Klk8 | 4887 | 0.061 | 0.1465 | No |
| 25 | Slc16a7 | 4965 | 0.057 | 0.1439 | No |
| 26 | Gpr19 | 4990 | 0.055 | 0.1455 | No |
| 27 | Brdt | 5387 | 0.028 | 0.1153 | No |
| 28 | Cdkal1 | 5443 | 0.024 | 0.1124 | No |
| 29 | Camk1d | 5557 | 0.017 | 0.1043 | No |
| 30 | Entpd7 | 5589 | 0.015 | 0.1028 | No |
| 31 | Itgb1bp2 | 5741 | 0.005 | 0.0909 | No |
| 32 | Zbtb16 | 5817 | 0.000 | 0.0848 | No |
| 33 | Asb7 | 6213 | -0.027 | 0.0546 | No |
| 34 | Mast3 | 6352 | -0.036 | 0.0458 | No |
| 35 | Slc25a23 | 6408 | -0.041 | 0.0439 | No |
| 36 | Mx2 | 6710 | -0.062 | 0.0235 | No |
| 37 | Bard1 | 6723 | -0.063 | 0.0265 | No |
| 38 | Thrb | 6792 | -0.068 | 0.0254 | No |
| 39 | Ybx2 | 6983 | -0.082 | 0.0152 | No |
| 40 | Tgfb2 | 7236 | -0.099 | 0.0011 | No |
| 41 | Skil | 7457 | -0.114 | -0.0094 | No |
| 42 | Nr6a1 | 7478 | -0.115 | -0.0037 | No |
| 43 | Btg2 | 7875 | -0.144 | -0.0265 | No |
| 44 | Cpeb3 | 7946 | -0.149 | -0.0226 | No |
| 45 | Snn | 8031 | -0.155 | -0.0196 | No |
| 46 | Ypel1 | 8079 | -0.158 | -0.0133 | No |
| 47 | Myo15a | 8205 | -0.166 | -0.0128 | No |
| 48 | Tent5c | 8724 | -0.202 | -0.0418 | No |
| 49 | Selenop | 8797 | -0.209 | -0.0343 | No |
| 50 | Kmt2d | 8993 | -0.224 | -0.0358 | No |
| 51 | Idua | 9803 | -0.289 | -0.0827 | No |
| 52 | Prkn | 9858 | -0.295 | -0.0683 | No |
| 53 | Coq8a | 10600 | -0.372 | -0.1044 | No |
| 54 | Lfng | 10870 | -0.405 | -0.1004 | No |
| 55 | Gp1ba | 11203 | -0.448 | -0.0987 | No |
| 56 | Pdk2 | 11646 | -0.525 | -0.1009 | No |
| 57 | Clstn3 | 11907 | -0.594 | -0.0841 | No |
| 58 | Prodh | 12144 | -0.693 | -0.0590 | No |
| 59 | Ccdc106 | 12438 | -1.302 | 0.0002 | No |