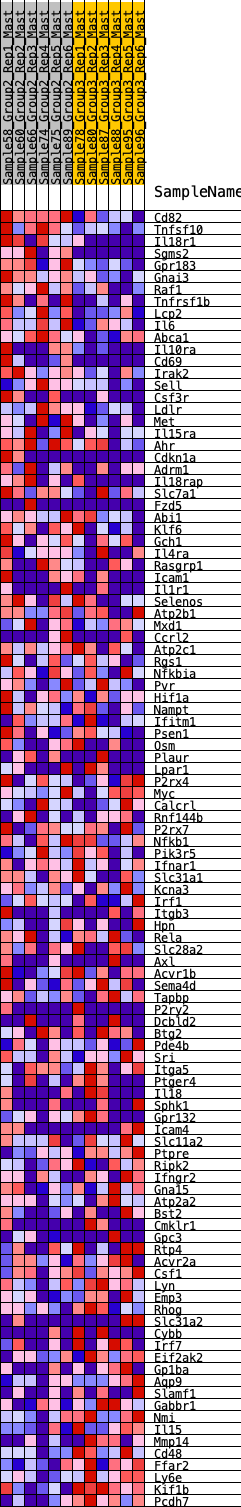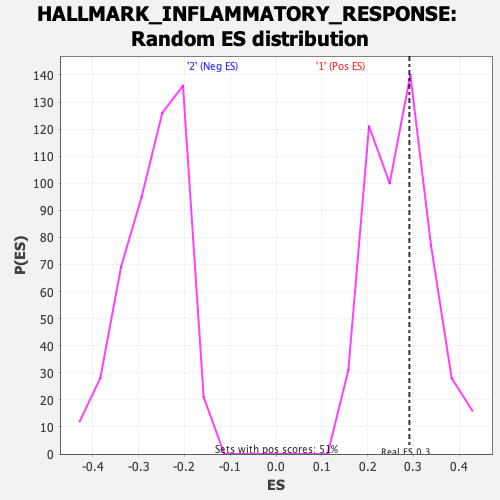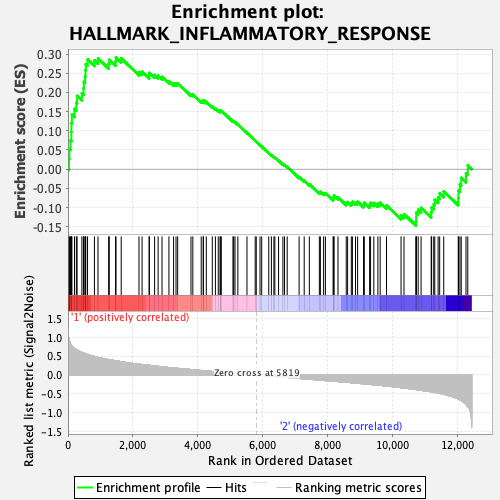
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.29133922 |
| Normalized Enrichment Score (NES) | 1.0764998 |
| Nominal p-value | 0.32163742 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd82 | 32 | 0.951 | 0.0261 | Yes |
| 2 | Tnfsf10 | 43 | 0.909 | 0.0527 | Yes |
| 3 | Il18r1 | 76 | 0.829 | 0.0751 | Yes |
| 4 | Sgms2 | 103 | 0.787 | 0.0967 | Yes |
| 5 | Gpr183 | 106 | 0.784 | 0.1202 | Yes |
| 6 | Gnai3 | 121 | 0.767 | 0.1422 | Yes |
| 7 | Raf1 | 200 | 0.708 | 0.1572 | Yes |
| 8 | Tnfrsf1b | 257 | 0.676 | 0.1730 | Yes |
| 9 | Lcp2 | 280 | 0.661 | 0.1912 | Yes |
| 10 | Il6 | 428 | 0.598 | 0.1973 | Yes |
| 11 | Abca1 | 479 | 0.584 | 0.2109 | Yes |
| 12 | Il10ra | 489 | 0.581 | 0.2277 | Yes |
| 13 | Cd69 | 526 | 0.566 | 0.2418 | Yes |
| 14 | Irak2 | 538 | 0.561 | 0.2578 | Yes |
| 15 | Sell | 548 | 0.559 | 0.2739 | Yes |
| 16 | Csf3r | 602 | 0.541 | 0.2860 | Yes |
| 17 | Ldlr | 814 | 0.489 | 0.2836 | Yes |
| 18 | Met | 924 | 0.465 | 0.2888 | Yes |
| 19 | Il15ra | 1253 | 0.408 | 0.2745 | Yes |
| 20 | Ahr | 1271 | 0.405 | 0.2853 | Yes |
| 21 | Cdkn1a | 1465 | 0.376 | 0.2810 | Yes |
| 22 | Adrm1 | 1478 | 0.374 | 0.2913 | Yes |
| 23 | Il18rap | 1639 | 0.352 | 0.2890 | No |
| 24 | Slc7a1 | 2187 | 0.284 | 0.2532 | No |
| 25 | Fzd5 | 2282 | 0.276 | 0.2539 | No |
| 26 | Abi1 | 2499 | 0.257 | 0.2441 | No |
| 27 | Klf6 | 2507 | 0.257 | 0.2513 | No |
| 28 | Gch1 | 2668 | 0.239 | 0.2455 | No |
| 29 | Il4ra | 2776 | 0.229 | 0.2438 | No |
| 30 | Rasgrp1 | 2897 | 0.216 | 0.2406 | No |
| 31 | Icam1 | 3110 | 0.197 | 0.2293 | No |
| 32 | Il1r1 | 3253 | 0.185 | 0.2234 | No |
| 33 | Selenos | 3328 | 0.179 | 0.2228 | No |
| 34 | Atp2b1 | 3379 | 0.176 | 0.2240 | No |
| 35 | Mxd1 | 3789 | 0.143 | 0.1952 | No |
| 36 | Ccrl2 | 3846 | 0.140 | 0.1949 | No |
| 37 | Atp2c1 | 4104 | 0.118 | 0.1776 | No |
| 38 | Rgs1 | 4168 | 0.114 | 0.1759 | No |
| 39 | Nfkbia | 4169 | 0.114 | 0.1794 | No |
| 40 | Pvr | 4261 | 0.108 | 0.1752 | No |
| 41 | Hif1a | 4445 | 0.092 | 0.1632 | No |
| 42 | Nampt | 4541 | 0.084 | 0.1580 | No |
| 43 | Ifitm1 | 4629 | 0.077 | 0.1532 | No |
| 44 | Psen1 | 4688 | 0.074 | 0.1508 | No |
| 45 | Osm | 4691 | 0.074 | 0.1528 | No |
| 46 | Plaur | 4723 | 0.072 | 0.1525 | No |
| 47 | Lpar1 | 5083 | 0.049 | 0.1248 | No |
| 48 | P2rx4 | 5104 | 0.047 | 0.1246 | No |
| 49 | Myc | 5145 | 0.044 | 0.1227 | No |
| 50 | Calcrl | 5233 | 0.038 | 0.1168 | No |
| 51 | Rnf144b | 5514 | 0.020 | 0.0947 | No |
| 52 | P2rx7 | 5767 | 0.003 | 0.0743 | No |
| 53 | Nfkb1 | 5800 | 0.002 | 0.0718 | No |
| 54 | Pik3r5 | 5916 | -0.006 | 0.0626 | No |
| 55 | Ifnar1 | 5968 | -0.010 | 0.0588 | No |
| 56 | Slc31a1 | 6185 | -0.025 | 0.0420 | No |
| 57 | Kcna3 | 6266 | -0.031 | 0.0365 | No |
| 58 | Irf1 | 6343 | -0.035 | 0.0314 | No |
| 59 | Itgb3 | 6373 | -0.037 | 0.0302 | No |
| 60 | Hpn | 6487 | -0.046 | 0.0224 | No |
| 61 | Rela | 6621 | -0.055 | 0.0132 | No |
| 62 | Slc28a2 | 6671 | -0.059 | 0.0110 | No |
| 63 | Axl | 6753 | -0.065 | 0.0064 | No |
| 64 | Acvr1b | 7120 | -0.091 | -0.0205 | No |
| 65 | Sema4d | 7277 | -0.101 | -0.0301 | No |
| 66 | Tapbp | 7435 | -0.112 | -0.0395 | No |
| 67 | P2ry2 | 7744 | -0.133 | -0.0604 | No |
| 68 | Dcbld2 | 7775 | -0.136 | -0.0588 | No |
| 69 | Btg2 | 7875 | -0.144 | -0.0625 | No |
| 70 | Pde4b | 7930 | -0.148 | -0.0624 | No |
| 71 | Sri | 8167 | -0.164 | -0.0766 | No |
| 72 | Itga5 | 8185 | -0.165 | -0.0730 | No |
| 73 | Ptger4 | 8197 | -0.165 | -0.0689 | No |
| 74 | Il18 | 8318 | -0.174 | -0.0734 | No |
| 75 | Sphk1 | 8572 | -0.191 | -0.0881 | No |
| 76 | Gpr132 | 8615 | -0.194 | -0.0857 | No |
| 77 | Icam4 | 8728 | -0.203 | -0.0886 | No |
| 78 | Slc11a2 | 8762 | -0.206 | -0.0851 | No |
| 79 | Ptpre | 8858 | -0.213 | -0.0864 | No |
| 80 | Ripk2 | 8923 | -0.218 | -0.0850 | No |
| 81 | Ifngr2 | 9105 | -0.233 | -0.0926 | No |
| 82 | Gna15 | 9129 | -0.234 | -0.0874 | No |
| 83 | Atp2a2 | 9293 | -0.248 | -0.0932 | No |
| 84 | Bst2 | 9322 | -0.251 | -0.0879 | No |
| 85 | Cmklr1 | 9422 | -0.259 | -0.0881 | No |
| 86 | Gpc3 | 9547 | -0.268 | -0.0901 | No |
| 87 | Rtp4 | 9617 | -0.273 | -0.0874 | No |
| 88 | Acvr2a | 9813 | -0.290 | -0.0945 | No |
| 89 | Csf1 | 10261 | -0.336 | -0.1206 | No |
| 90 | Lyn | 10352 | -0.345 | -0.1175 | No |
| 91 | Emp3 | 10719 | -0.386 | -0.1356 | No |
| 92 | Rhog | 10730 | -0.387 | -0.1247 | No |
| 93 | Slc31a2 | 10731 | -0.387 | -0.1130 | No |
| 94 | Cybb | 10788 | -0.393 | -0.1057 | No |
| 95 | Irf7 | 10878 | -0.406 | -0.1007 | No |
| 96 | Eif2ak2 | 11189 | -0.445 | -0.1124 | No |
| 97 | Gp1ba | 11203 | -0.448 | -0.1000 | No |
| 98 | Aqp9 | 11268 | -0.457 | -0.0914 | No |
| 99 | Slamf1 | 11300 | -0.463 | -0.0800 | No |
| 100 | Gabbr1 | 11403 | -0.477 | -0.0738 | No |
| 101 | Nmi | 11451 | -0.485 | -0.0630 | No |
| 102 | Il15 | 11576 | -0.506 | -0.0578 | No |
| 103 | Mmp14 | 12023 | -0.634 | -0.0748 | No |
| 104 | Cd48 | 12031 | -0.638 | -0.0562 | No |
| 105 | Ffar2 | 12075 | -0.659 | -0.0398 | No |
| 106 | Ly6e | 12115 | -0.676 | -0.0226 | No |
| 107 | Kif1b | 12262 | -0.779 | -0.0109 | No |
| 108 | Pcdh7 | 12317 | -0.840 | 0.0101 | No |