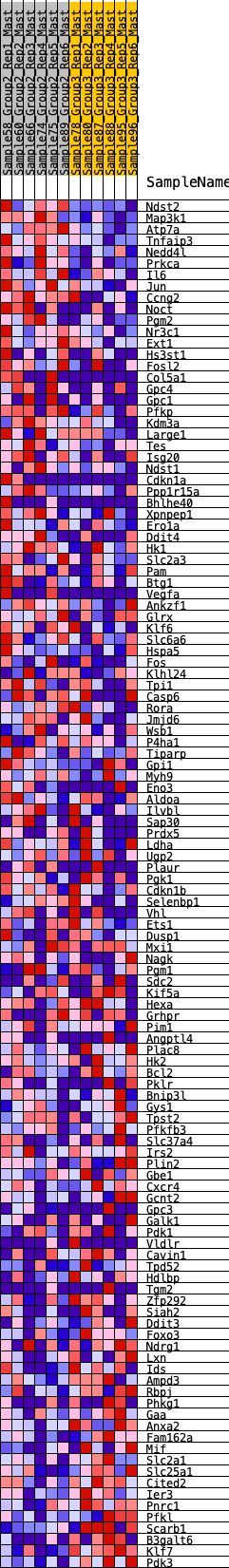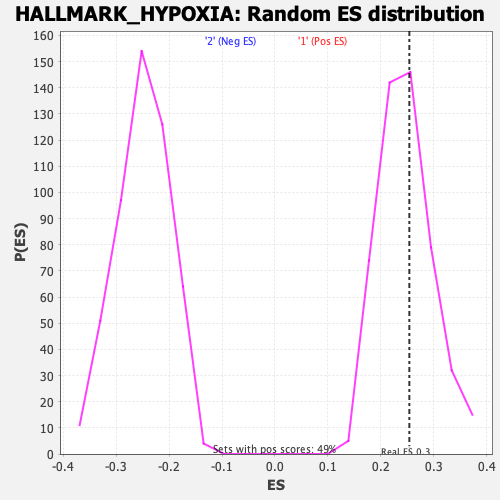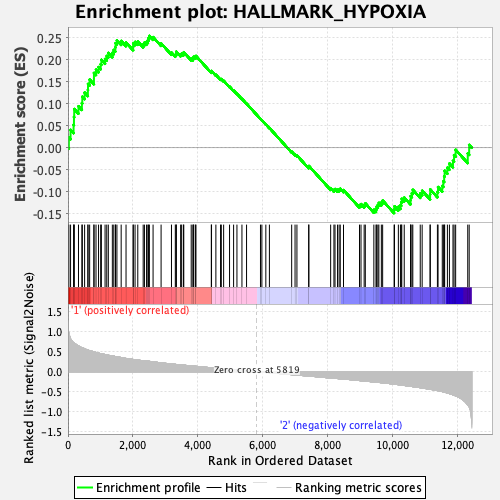
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.25371984 |
| Normalized Enrichment Score (NES) | 1.0268612 |
| Nominal p-value | 0.40567952 |
| FDR q-value | 0.7812424 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ndst2 | 27 | 0.975 | 0.0232 | Yes |
| 2 | Map3k1 | 79 | 0.824 | 0.0406 | Yes |
| 3 | Atp7a | 172 | 0.724 | 0.0520 | Yes |
| 4 | Tnfaip3 | 184 | 0.717 | 0.0699 | Yes |
| 5 | Nedd4l | 192 | 0.713 | 0.0879 | Yes |
| 6 | Prkca | 323 | 0.643 | 0.0942 | Yes |
| 7 | Il6 | 428 | 0.598 | 0.1013 | Yes |
| 8 | Jun | 441 | 0.595 | 0.1159 | Yes |
| 9 | Ccng2 | 512 | 0.573 | 0.1252 | Yes |
| 10 | Noct | 612 | 0.538 | 0.1312 | Yes |
| 11 | Pgm2 | 617 | 0.537 | 0.1449 | Yes |
| 12 | Nr3c1 | 667 | 0.526 | 0.1546 | Yes |
| 13 | Ext1 | 799 | 0.494 | 0.1569 | Yes |
| 14 | Hs3st1 | 800 | 0.494 | 0.1698 | Yes |
| 15 | Fosl2 | 863 | 0.478 | 0.1772 | Yes |
| 16 | Col5a1 | 941 | 0.462 | 0.1830 | Yes |
| 17 | Gpc4 | 1007 | 0.449 | 0.1895 | Yes |
| 18 | Gpc1 | 1028 | 0.445 | 0.1995 | Yes |
| 19 | Pfkp | 1142 | 0.427 | 0.2014 | Yes |
| 20 | Kdm3a | 1194 | 0.419 | 0.2082 | Yes |
| 21 | Large1 | 1242 | 0.409 | 0.2151 | Yes |
| 22 | Tes | 1366 | 0.391 | 0.2153 | Yes |
| 23 | Isg20 | 1410 | 0.386 | 0.2219 | Yes |
| 24 | Ndst1 | 1459 | 0.378 | 0.2279 | Yes |
| 25 | Cdkn1a | 1465 | 0.376 | 0.2373 | Yes |
| 26 | Ppp1r15a | 1507 | 0.370 | 0.2436 | Yes |
| 27 | Bhlhe40 | 1638 | 0.352 | 0.2423 | Yes |
| 28 | Xpnpep1 | 1788 | 0.330 | 0.2388 | Yes |
| 29 | Ero1a | 2007 | 0.304 | 0.2290 | Yes |
| 30 | Ddit4 | 2010 | 0.304 | 0.2368 | Yes |
| 31 | Hk1 | 2066 | 0.297 | 0.2401 | Yes |
| 32 | Slc2a3 | 2150 | 0.288 | 0.2409 | Yes |
| 33 | Pam | 2316 | 0.273 | 0.2346 | Yes |
| 34 | Btg1 | 2355 | 0.269 | 0.2386 | Yes |
| 35 | Vegfa | 2420 | 0.264 | 0.2403 | Yes |
| 36 | Ankzf1 | 2461 | 0.261 | 0.2438 | Yes |
| 37 | Glrx | 2482 | 0.259 | 0.2490 | Yes |
| 38 | Klf6 | 2507 | 0.257 | 0.2537 | Yes |
| 39 | Slc6a6 | 2621 | 0.244 | 0.2509 | No |
| 40 | Hspa5 | 2868 | 0.219 | 0.2367 | No |
| 41 | Fos | 3188 | 0.190 | 0.2158 | No |
| 42 | Klhl24 | 3306 | 0.181 | 0.2110 | No |
| 43 | Tpi1 | 3333 | 0.179 | 0.2135 | No |
| 44 | Casp6 | 3336 | 0.178 | 0.2180 | No |
| 45 | Rora | 3470 | 0.167 | 0.2116 | No |
| 46 | Jmjd6 | 3501 | 0.165 | 0.2135 | No |
| 47 | Wsb1 | 3556 | 0.160 | 0.2133 | No |
| 48 | P4ha1 | 3567 | 0.160 | 0.2166 | No |
| 49 | Tiparp | 3796 | 0.143 | 0.2019 | No |
| 50 | Gpi1 | 3848 | 0.139 | 0.2014 | No |
| 51 | Myh9 | 3861 | 0.138 | 0.2040 | No |
| 52 | Eno3 | 3875 | 0.137 | 0.2065 | No |
| 53 | Aldoa | 3932 | 0.132 | 0.2054 | No |
| 54 | Ilvbl | 3940 | 0.131 | 0.2083 | No |
| 55 | Sap30 | 4415 | 0.094 | 0.1723 | No |
| 56 | Prdx5 | 4422 | 0.093 | 0.1742 | No |
| 57 | Ldha | 4556 | 0.082 | 0.1655 | No |
| 58 | Ugp2 | 4700 | 0.073 | 0.1559 | No |
| 59 | Plaur | 4723 | 0.072 | 0.1559 | No |
| 60 | Pgk1 | 4795 | 0.068 | 0.1520 | No |
| 61 | Cdkn1b | 4977 | 0.056 | 0.1387 | No |
| 62 | Selenbp1 | 5102 | 0.047 | 0.1299 | No |
| 63 | Vhl | 5205 | 0.039 | 0.1226 | No |
| 64 | Ets1 | 5359 | 0.030 | 0.1110 | No |
| 65 | Dusp1 | 5501 | 0.021 | 0.1001 | No |
| 66 | Mxi1 | 5930 | -0.006 | 0.0655 | No |
| 67 | Nagk | 5973 | -0.010 | 0.0624 | No |
| 68 | Pgm1 | 6095 | -0.019 | 0.0531 | No |
| 69 | Sdc2 | 6205 | -0.026 | 0.0449 | No |
| 70 | Kif5a | 6891 | -0.075 | -0.0087 | No |
| 71 | Hexa | 6995 | -0.083 | -0.0149 | No |
| 72 | Grhpr | 7053 | -0.087 | -0.0173 | No |
| 73 | Pim1 | 7415 | -0.111 | -0.0437 | No |
| 74 | Angptl4 | 7421 | -0.111 | -0.0412 | No |
| 75 | Plac8 | 8092 | -0.159 | -0.0914 | No |
| 76 | Hk2 | 8193 | -0.165 | -0.0952 | No |
| 77 | Bcl2 | 8226 | -0.168 | -0.0934 | No |
| 78 | Pklr | 8304 | -0.173 | -0.0952 | No |
| 79 | Bnip3l | 8352 | -0.176 | -0.0944 | No |
| 80 | Gys1 | 8388 | -0.178 | -0.0926 | No |
| 81 | Tpst2 | 8488 | -0.185 | -0.0958 | No |
| 82 | Pfkfb3 | 8981 | -0.224 | -0.1299 | No |
| 83 | Slc37a4 | 9027 | -0.227 | -0.1276 | No |
| 84 | Irs2 | 9124 | -0.234 | -0.1293 | No |
| 85 | Plin2 | 9160 | -0.237 | -0.1259 | No |
| 86 | Gbe1 | 9423 | -0.259 | -0.1404 | No |
| 87 | Cxcr4 | 9489 | -0.263 | -0.1389 | No |
| 88 | Gcnt2 | 9509 | -0.266 | -0.1335 | No |
| 89 | Gpc3 | 9547 | -0.268 | -0.1295 | No |
| 90 | Galk1 | 9577 | -0.270 | -0.1248 | No |
| 91 | Pdk1 | 9654 | -0.276 | -0.1237 | No |
| 92 | Vldlr | 9694 | -0.280 | -0.1196 | No |
| 93 | Cavin1 | 10047 | -0.314 | -0.1400 | No |
| 94 | Tpd52 | 10059 | -0.315 | -0.1326 | No |
| 95 | Hdlbp | 10178 | -0.327 | -0.1337 | No |
| 96 | Tgm2 | 10242 | -0.333 | -0.1301 | No |
| 97 | Zfp292 | 10266 | -0.336 | -0.1232 | No |
| 98 | Siah2 | 10283 | -0.337 | -0.1157 | No |
| 99 | Ddit3 | 10359 | -0.346 | -0.1127 | No |
| 100 | Foxo3 | 10553 | -0.366 | -0.1188 | No |
| 101 | Ndrg1 | 10556 | -0.366 | -0.1094 | No |
| 102 | Lxn | 10595 | -0.372 | -0.1028 | No |
| 103 | Ids | 10623 | -0.375 | -0.0952 | No |
| 104 | Ampd3 | 10852 | -0.402 | -0.1032 | No |
| 105 | Rbpj | 10911 | -0.409 | -0.0972 | No |
| 106 | Phkg1 | 11156 | -0.439 | -0.1056 | No |
| 107 | Gaa | 11160 | -0.440 | -0.0943 | No |
| 108 | Anxa2 | 11381 | -0.474 | -0.0998 | No |
| 109 | Fam162a | 11401 | -0.477 | -0.0889 | No |
| 110 | Mif | 11531 | -0.499 | -0.0863 | No |
| 111 | Slc2a1 | 11564 | -0.504 | -0.0758 | No |
| 112 | Slc25a1 | 11590 | -0.511 | -0.0645 | No |
| 113 | Cited2 | 11604 | -0.514 | -0.0521 | No |
| 114 | Ier3 | 11689 | -0.534 | -0.0450 | No |
| 115 | Pnrc1 | 11751 | -0.550 | -0.0356 | No |
| 116 | Pfkl | 11861 | -0.580 | -0.0293 | No |
| 117 | Scarb1 | 11894 | -0.590 | -0.0165 | No |
| 118 | B3galt6 | 11943 | -0.605 | -0.0046 | No |
| 119 | Klf7 | 12316 | -0.839 | -0.0129 | No |
| 120 | Pdk3 | 12360 | -0.878 | 0.0066 | No |