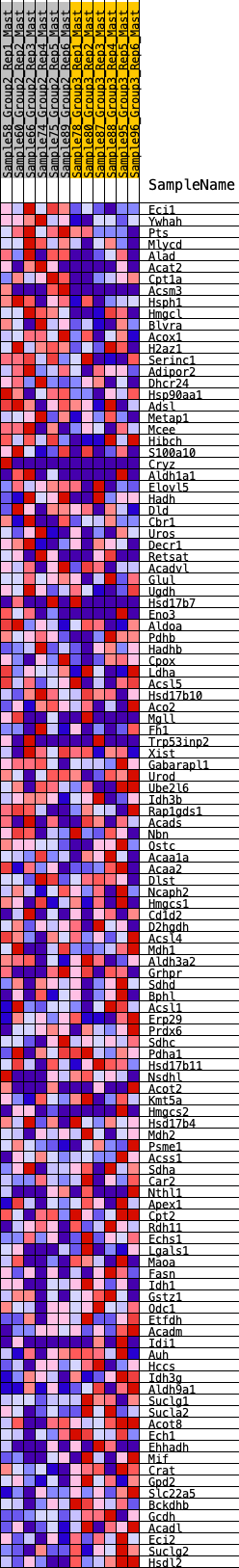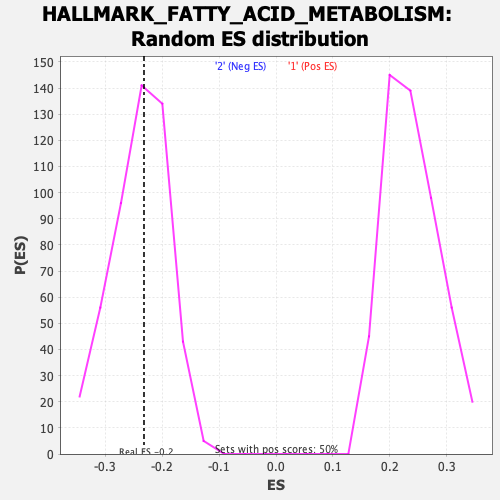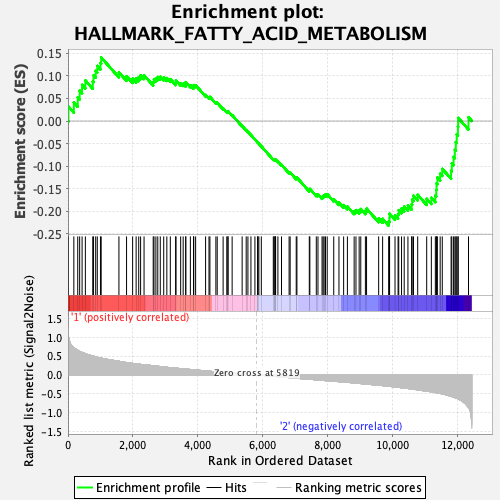
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23169047 |
| Normalized Enrichment Score (NES) | -0.96756226 |
| Nominal p-value | 0.52112675 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eci1 | 15 | 1.043 | 0.0318 | No |
| 2 | Ywhah | 180 | 0.718 | 0.0412 | No |
| 3 | Pts | 300 | 0.652 | 0.0522 | No |
| 4 | Mlycd | 359 | 0.626 | 0.0673 | No |
| 5 | Alad | 434 | 0.597 | 0.0802 | No |
| 6 | Acat2 | 535 | 0.562 | 0.0898 | No |
| 7 | Cpt1a | 763 | 0.503 | 0.0874 | No |
| 8 | Acsm3 | 787 | 0.496 | 0.1012 | No |
| 9 | Hsph1 | 853 | 0.480 | 0.1111 | No |
| 10 | Hmgcl | 902 | 0.469 | 0.1221 | No |
| 11 | Blvra | 1000 | 0.451 | 0.1284 | No |
| 12 | Acox1 | 1022 | 0.446 | 0.1409 | No |
| 13 | H2az1 | 1569 | 0.361 | 0.1080 | No |
| 14 | Serinc1 | 1804 | 0.327 | 0.0993 | No |
| 15 | Adipor2 | 1991 | 0.306 | 0.0939 | No |
| 16 | Dhcr24 | 2099 | 0.294 | 0.0946 | No |
| 17 | Hsp90aa1 | 2179 | 0.285 | 0.0972 | No |
| 18 | Adsl | 2237 | 0.279 | 0.1014 | No |
| 19 | Metap1 | 2343 | 0.270 | 0.1014 | No |
| 20 | Mcee | 2625 | 0.244 | 0.0863 | No |
| 21 | Hibch | 2649 | 0.241 | 0.0921 | No |
| 22 | S100a10 | 2708 | 0.236 | 0.0948 | No |
| 23 | Cryz | 2766 | 0.229 | 0.0974 | No |
| 24 | Aldh1a1 | 2843 | 0.222 | 0.0983 | No |
| 25 | Elovl5 | 2949 | 0.211 | 0.0965 | No |
| 26 | Hadh | 3045 | 0.203 | 0.0952 | No |
| 27 | Dld | 3152 | 0.194 | 0.0927 | No |
| 28 | Cbr1 | 3316 | 0.180 | 0.0852 | No |
| 29 | Uros | 3327 | 0.179 | 0.0900 | No |
| 30 | Decr1 | 3471 | 0.167 | 0.0837 | No |
| 31 | Retsat | 3548 | 0.161 | 0.0827 | No |
| 32 | Acadvl | 3627 | 0.156 | 0.0813 | No |
| 33 | Glul | 3628 | 0.155 | 0.0862 | No |
| 34 | Ugdh | 3770 | 0.144 | 0.0793 | No |
| 35 | Hsd17b7 | 3866 | 0.138 | 0.0760 | No |
| 36 | Eno3 | 3875 | 0.137 | 0.0796 | No |
| 37 | Aldoa | 3932 | 0.132 | 0.0793 | No |
| 38 | Pdhb | 4239 | 0.109 | 0.0579 | No |
| 39 | Hadhb | 4344 | 0.100 | 0.0526 | No |
| 40 | Cpox | 4372 | 0.098 | 0.0535 | No |
| 41 | Ldha | 4556 | 0.082 | 0.0413 | No |
| 42 | Acsl5 | 4597 | 0.079 | 0.0405 | No |
| 43 | Hsd17b10 | 4779 | 0.069 | 0.0280 | No |
| 44 | Aco2 | 4892 | 0.061 | 0.0209 | No |
| 45 | Mgll | 4928 | 0.059 | 0.0199 | No |
| 46 | Fh1 | 4929 | 0.059 | 0.0218 | No |
| 47 | Trp53inp2 | 5057 | 0.050 | 0.0131 | No |
| 48 | Xist | 5365 | 0.030 | -0.0109 | No |
| 49 | Gabarapl1 | 5490 | 0.021 | -0.0203 | No |
| 50 | Urod | 5543 | 0.018 | -0.0239 | No |
| 51 | Ube2l6 | 5636 | 0.012 | -0.0310 | No |
| 52 | Idh3b | 5759 | 0.003 | -0.0408 | No |
| 53 | Rap1gds1 | 5840 | -0.001 | -0.0473 | No |
| 54 | Acads | 5851 | -0.001 | -0.0481 | No |
| 55 | Nbn | 5880 | -0.003 | -0.0502 | No |
| 56 | Ostc | 5956 | -0.009 | -0.0560 | No |
| 57 | Acaa1a | 6325 | -0.034 | -0.0848 | No |
| 58 | Acaa2 | 6348 | -0.035 | -0.0855 | No |
| 59 | Dlst | 6371 | -0.037 | -0.0861 | No |
| 60 | Ncaph2 | 6392 | -0.040 | -0.0865 | No |
| 61 | Hmgcs1 | 6396 | -0.040 | -0.0855 | No |
| 62 | Cd1d2 | 6467 | -0.044 | -0.0897 | No |
| 63 | D2hgdh | 6578 | -0.053 | -0.0970 | No |
| 64 | Acsl4 | 6812 | -0.069 | -0.1137 | No |
| 65 | Mdh1 | 6844 | -0.072 | -0.1140 | No |
| 66 | Aldh3a2 | 7031 | -0.085 | -0.1264 | No |
| 67 | Grhpr | 7053 | -0.087 | -0.1253 | No |
| 68 | Sdhd | 7434 | -0.112 | -0.1526 | No |
| 69 | Bphl | 7451 | -0.113 | -0.1503 | No |
| 70 | Acsl1 | 7650 | -0.126 | -0.1624 | No |
| 71 | Erp29 | 7701 | -0.130 | -0.1623 | No |
| 72 | Prdx6 | 7827 | -0.140 | -0.1680 | No |
| 73 | Sdhc | 7859 | -0.142 | -0.1661 | No |
| 74 | Pdha1 | 7891 | -0.144 | -0.1640 | No |
| 75 | Hsd17b11 | 7933 | -0.148 | -0.1627 | No |
| 76 | Nsdhl | 7986 | -0.152 | -0.1621 | No |
| 77 | Acot2 | 8187 | -0.165 | -0.1731 | No |
| 78 | Kmt5a | 8348 | -0.176 | -0.1805 | No |
| 79 | Hmgcs2 | 8492 | -0.185 | -0.1862 | No |
| 80 | Hsd17b4 | 8608 | -0.193 | -0.1895 | No |
| 81 | Mdh2 | 8816 | -0.211 | -0.1996 | No |
| 82 | Psme1 | 8872 | -0.214 | -0.1973 | No |
| 83 | Acss1 | 8970 | -0.223 | -0.1981 | No |
| 84 | Sdha | 9020 | -0.227 | -0.1949 | No |
| 85 | Car2 | 9162 | -0.237 | -0.1988 | No |
| 86 | Nthl1 | 9198 | -0.240 | -0.1941 | No |
| 87 | Apex1 | 9571 | -0.270 | -0.2157 | No |
| 88 | Cpt2 | 9693 | -0.280 | -0.2167 | No |
| 89 | Rdh11 | 9879 | -0.296 | -0.2223 | Yes |
| 90 | Echs1 | 9906 | -0.300 | -0.2149 | Yes |
| 91 | Lgals1 | 9907 | -0.300 | -0.2055 | Yes |
| 92 | Maoa | 10075 | -0.316 | -0.2090 | Yes |
| 93 | Fasn | 10169 | -0.326 | -0.2062 | Yes |
| 94 | Idh1 | 10190 | -0.328 | -0.1975 | Yes |
| 95 | Gstz1 | 10276 | -0.337 | -0.1937 | Yes |
| 96 | Odc1 | 10358 | -0.346 | -0.1893 | Yes |
| 97 | Etfdh | 10473 | -0.358 | -0.1873 | Yes |
| 98 | Acadm | 10586 | -0.370 | -0.1846 | Yes |
| 99 | Idi1 | 10603 | -0.373 | -0.1741 | Yes |
| 100 | Auh | 10645 | -0.378 | -0.1655 | Yes |
| 101 | Hccs | 10774 | -0.391 | -0.1635 | Yes |
| 102 | Idh3g | 11051 | -0.427 | -0.1724 | Yes |
| 103 | Aldh9a1 | 11194 | -0.446 | -0.1698 | Yes |
| 104 | Suclg1 | 11317 | -0.465 | -0.1650 | Yes |
| 105 | Sucla2 | 11347 | -0.470 | -0.1525 | Yes |
| 106 | Acot8 | 11355 | -0.471 | -0.1382 | Yes |
| 107 | Ech1 | 11380 | -0.474 | -0.1251 | Yes |
| 108 | Ehhadh | 11466 | -0.487 | -0.1166 | Yes |
| 109 | Mif | 11531 | -0.499 | -0.1060 | Yes |
| 110 | Crat | 11803 | -0.563 | -0.1102 | Yes |
| 111 | Gpd2 | 11825 | -0.570 | -0.0938 | Yes |
| 112 | Slc22a5 | 11879 | -0.587 | -0.0795 | Yes |
| 113 | Bckdhb | 11919 | -0.599 | -0.0638 | Yes |
| 114 | Gcdh | 11947 | -0.606 | -0.0468 | Yes |
| 115 | Acadl | 11976 | -0.615 | -0.0296 | Yes |
| 116 | Eci2 | 12013 | -0.631 | -0.0125 | Yes |
| 117 | Suclg2 | 12021 | -0.633 | 0.0069 | Yes |
| 118 | Hsdl2 | 12339 | -0.856 | 0.0083 | Yes |