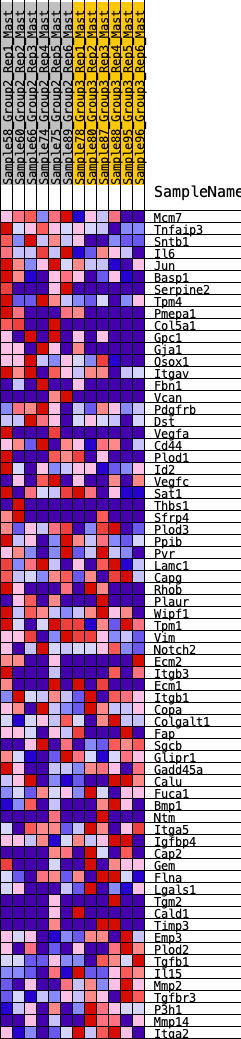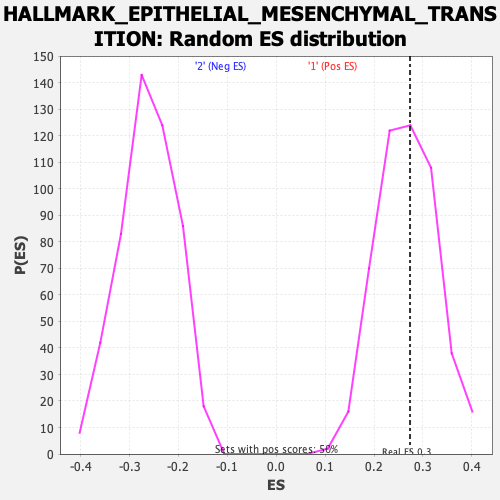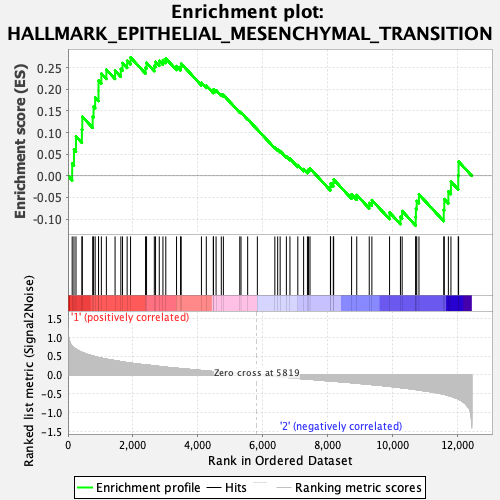
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.27353248 |
| Normalized Enrichment Score (NES) | 1.028808 |
| Nominal p-value | 0.43548387 |
| FDR q-value | 0.8399985 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mcm7 | 129 | 0.758 | 0.0285 | Yes |
| 2 | Tnfaip3 | 184 | 0.717 | 0.0609 | Yes |
| 3 | Sntb1 | 246 | 0.683 | 0.0910 | Yes |
| 4 | Il6 | 428 | 0.598 | 0.1071 | Yes |
| 5 | Jun | 441 | 0.595 | 0.1367 | Yes |
| 6 | Basp1 | 761 | 0.504 | 0.1367 | Yes |
| 7 | Serpine2 | 788 | 0.496 | 0.1600 | Yes |
| 8 | Tpm4 | 837 | 0.484 | 0.1810 | Yes |
| 9 | Pmepa1 | 940 | 0.462 | 0.1965 | Yes |
| 10 | Col5a1 | 941 | 0.462 | 0.2202 | Yes |
| 11 | Gpc1 | 1028 | 0.445 | 0.2361 | Yes |
| 12 | Gja1 | 1182 | 0.421 | 0.2453 | Yes |
| 13 | Qsox1 | 1450 | 0.379 | 0.2432 | Yes |
| 14 | Itgav | 1628 | 0.354 | 0.2470 | Yes |
| 15 | Fbn1 | 1680 | 0.345 | 0.2606 | Yes |
| 16 | Vcan | 1822 | 0.325 | 0.2659 | Yes |
| 17 | Pdgfrb | 1927 | 0.313 | 0.2735 | Yes |
| 18 | Dst | 2389 | 0.267 | 0.2500 | No |
| 19 | Vegfa | 2420 | 0.264 | 0.2611 | No |
| 20 | Cd44 | 2659 | 0.239 | 0.2541 | No |
| 21 | Plod1 | 2697 | 0.237 | 0.2633 | No |
| 22 | Id2 | 2812 | 0.225 | 0.2656 | No |
| 23 | Vegfc | 2925 | 0.213 | 0.2675 | No |
| 24 | Sat1 | 3013 | 0.206 | 0.2711 | No |
| 25 | Thbs1 | 3344 | 0.178 | 0.2535 | No |
| 26 | Sfrp4 | 3467 | 0.167 | 0.2522 | No |
| 27 | Plod3 | 3487 | 0.167 | 0.2593 | No |
| 28 | Ppib | 4111 | 0.118 | 0.2149 | No |
| 29 | Pvr | 4261 | 0.108 | 0.2084 | No |
| 30 | Lamc1 | 4476 | 0.089 | 0.1957 | No |
| 31 | Capg | 4483 | 0.088 | 0.1998 | No |
| 32 | Rhob | 4562 | 0.081 | 0.1976 | No |
| 33 | Plaur | 4723 | 0.072 | 0.1884 | No |
| 34 | Wipf1 | 4787 | 0.068 | 0.1868 | No |
| 35 | Tpm1 | 5292 | 0.035 | 0.1479 | No |
| 36 | Vim | 5330 | 0.031 | 0.1465 | No |
| 37 | Notch2 | 5535 | 0.018 | 0.1309 | No |
| 38 | Ecm2 | 5834 | -0.000 | 0.1069 | No |
| 39 | Itgb3 | 6373 | -0.037 | 0.0653 | No |
| 40 | Ecm1 | 6460 | -0.044 | 0.0606 | No |
| 41 | Itgb1 | 6536 | -0.050 | 0.0571 | No |
| 42 | Copa | 6728 | -0.063 | 0.0449 | No |
| 43 | Colgalt1 | 6838 | -0.071 | 0.0397 | No |
| 44 | Fap | 7080 | -0.089 | 0.0249 | No |
| 45 | Sgcb | 7259 | -0.100 | 0.0156 | No |
| 46 | Glipr1 | 7377 | -0.108 | 0.0117 | No |
| 47 | Gadd45a | 7411 | -0.111 | 0.0147 | No |
| 48 | Calu | 7456 | -0.114 | 0.0170 | No |
| 49 | Fuca1 | 8085 | -0.158 | -0.0257 | No |
| 50 | Bmp1 | 8090 | -0.159 | -0.0178 | No |
| 51 | Ntm | 8174 | -0.164 | -0.0161 | No |
| 52 | Itga5 | 8185 | -0.165 | -0.0085 | No |
| 53 | Igfbp4 | 8737 | -0.204 | -0.0426 | No |
| 54 | Cap2 | 8896 | -0.216 | -0.0443 | No |
| 55 | Gem | 9281 | -0.246 | -0.0627 | No |
| 56 | Flna | 9362 | -0.254 | -0.0561 | No |
| 57 | Lgals1 | 9907 | -0.300 | -0.0847 | No |
| 58 | Tgm2 | 10242 | -0.333 | -0.0946 | No |
| 59 | Cald1 | 10295 | -0.339 | -0.0814 | No |
| 60 | Timp3 | 10712 | -0.385 | -0.0953 | No |
| 61 | Emp3 | 10719 | -0.386 | -0.0760 | No |
| 62 | Plod2 | 10744 | -0.388 | -0.0580 | No |
| 63 | Tgfb1 | 10813 | -0.397 | -0.0431 | No |
| 64 | Il15 | 11576 | -0.506 | -0.0787 | No |
| 65 | Mmp2 | 11593 | -0.512 | -0.0538 | No |
| 66 | Tgfbr3 | 11719 | -0.543 | -0.0360 | No |
| 67 | P3h1 | 11798 | -0.562 | -0.0135 | No |
| 68 | Mmp14 | 12023 | -0.634 | 0.0009 | No |
| 69 | Itga2 | 12034 | -0.638 | 0.0329 | No |