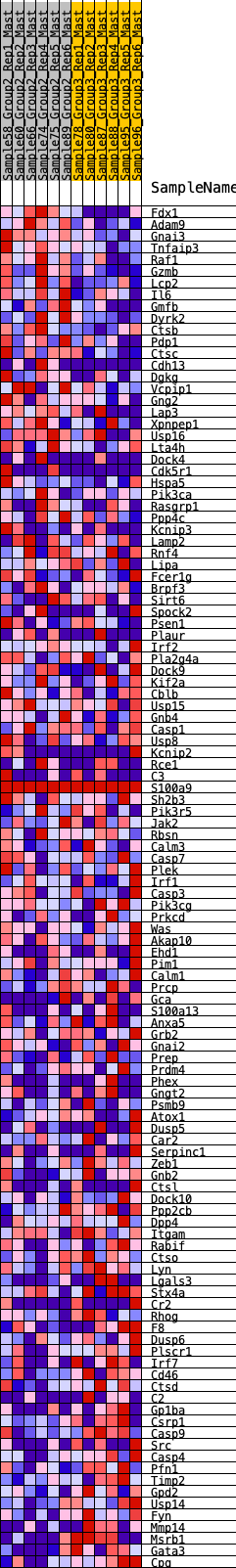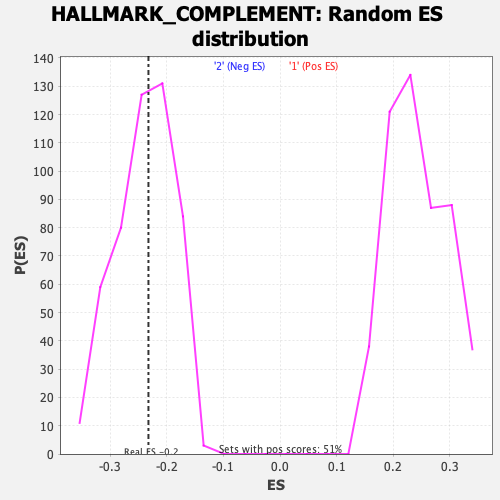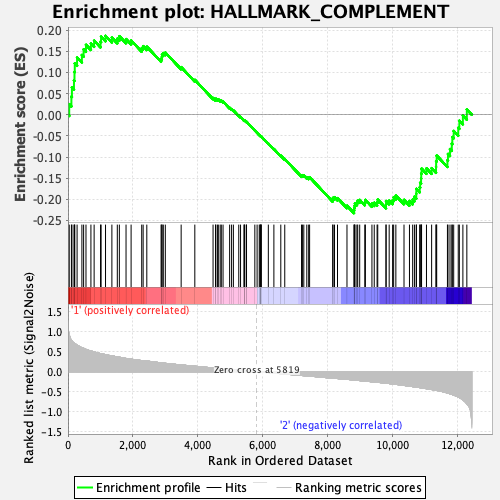
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.23270425 |
| Normalized Enrichment Score (NES) | -0.97166467 |
| Nominal p-value | 0.4969697 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdx1 | 38 | 0.934 | 0.0248 | No |
| 2 | Adam9 | 105 | 0.786 | 0.0429 | No |
| 3 | Gnai3 | 121 | 0.767 | 0.0646 | No |
| 4 | Tnfaip3 | 184 | 0.717 | 0.0810 | No |
| 5 | Raf1 | 200 | 0.708 | 0.1009 | No |
| 6 | Gzmb | 209 | 0.703 | 0.1213 | No |
| 7 | Lcp2 | 280 | 0.661 | 0.1353 | No |
| 8 | Il6 | 428 | 0.598 | 0.1413 | No |
| 9 | Gmfb | 481 | 0.583 | 0.1545 | No |
| 10 | Dyrk2 | 554 | 0.557 | 0.1653 | No |
| 11 | Ctsb | 706 | 0.516 | 0.1684 | No |
| 12 | Pdp1 | 805 | 0.492 | 0.1752 | No |
| 13 | Ctsc | 1003 | 0.450 | 0.1726 | No |
| 14 | Cdh13 | 1019 | 0.448 | 0.1848 | No |
| 15 | Dgkg | 1156 | 0.424 | 0.1864 | No |
| 16 | Vcpip1 | 1352 | 0.393 | 0.1823 | No |
| 17 | Gng2 | 1519 | 0.368 | 0.1799 | No |
| 18 | Lap3 | 1582 | 0.360 | 0.1856 | No |
| 19 | Xpnpep1 | 1788 | 0.330 | 0.1788 | No |
| 20 | Usp16 | 1944 | 0.312 | 0.1755 | No |
| 21 | Lta4h | 2271 | 0.277 | 0.1573 | No |
| 22 | Dock4 | 2315 | 0.273 | 0.1620 | No |
| 23 | Cdk5r1 | 2428 | 0.264 | 0.1608 | No |
| 24 | Hspa5 | 2868 | 0.219 | 0.1318 | No |
| 25 | Pik3ca | 2891 | 0.216 | 0.1364 | No |
| 26 | Rasgrp1 | 2897 | 0.216 | 0.1425 | No |
| 27 | Ppp4c | 2936 | 0.212 | 0.1457 | No |
| 28 | Kcnip3 | 2998 | 0.207 | 0.1470 | No |
| 29 | Lamp2 | 3486 | 0.167 | 0.1124 | No |
| 30 | Rnf4 | 3907 | 0.134 | 0.0823 | No |
| 31 | Lipa | 4469 | 0.090 | 0.0395 | No |
| 32 | Fcer1g | 4546 | 0.083 | 0.0358 | No |
| 33 | Brpf3 | 4553 | 0.082 | 0.0378 | No |
| 34 | Sirt6 | 4603 | 0.079 | 0.0362 | No |
| 35 | Spock2 | 4631 | 0.077 | 0.0363 | No |
| 36 | Psen1 | 4688 | 0.074 | 0.0339 | No |
| 37 | Plaur | 4723 | 0.072 | 0.0333 | No |
| 38 | Irf2 | 4777 | 0.069 | 0.0311 | No |
| 39 | Pla2g4a | 4980 | 0.055 | 0.0163 | No |
| 40 | Dock9 | 5040 | 0.051 | 0.0131 | No |
| 41 | Kif2a | 5095 | 0.047 | 0.0101 | No |
| 42 | Cblb | 5261 | 0.036 | -0.0022 | No |
| 43 | Usp15 | 5316 | 0.033 | -0.0056 | No |
| 44 | Gnb4 | 5425 | 0.026 | -0.0136 | No |
| 45 | Casp1 | 5429 | 0.025 | -0.0131 | No |
| 46 | Usp8 | 5442 | 0.024 | -0.0134 | No |
| 47 | Kcnip2 | 5493 | 0.021 | -0.0168 | No |
| 48 | Rce1 | 5500 | 0.021 | -0.0167 | No |
| 49 | C3 | 5756 | 0.004 | -0.0372 | No |
| 50 | S100a9 | 5826 | 0.000 | -0.0428 | No |
| 51 | Sh2b3 | 5892 | -0.004 | -0.0480 | No |
| 52 | Pik3r5 | 5916 | -0.006 | -0.0497 | No |
| 53 | Jak2 | 5940 | -0.007 | -0.0513 | No |
| 54 | Rbsn | 5942 | -0.007 | -0.0512 | No |
| 55 | Calm3 | 5944 | -0.008 | -0.0510 | No |
| 56 | Casp7 | 5950 | -0.008 | -0.0512 | No |
| 57 | Plek | 6172 | -0.024 | -0.0684 | No |
| 58 | Irf1 | 6343 | -0.035 | -0.0812 | No |
| 59 | Casp3 | 6556 | -0.051 | -0.0968 | No |
| 60 | Pik3cg | 6678 | -0.059 | -0.1049 | No |
| 61 | Prkcd | 7195 | -0.097 | -0.1439 | No |
| 62 | Was | 7222 | -0.098 | -0.1430 | No |
| 63 | Akap10 | 7265 | -0.100 | -0.1435 | No |
| 64 | Ehd1 | 7357 | -0.107 | -0.1476 | No |
| 65 | Pim1 | 7415 | -0.111 | -0.1489 | No |
| 66 | Calm1 | 7444 | -0.113 | -0.1479 | No |
| 67 | Prcp | 8155 | -0.163 | -0.2006 | No |
| 68 | Gca | 8165 | -0.164 | -0.1964 | No |
| 69 | S100a13 | 8211 | -0.167 | -0.1951 | No |
| 70 | Anxa5 | 8303 | -0.173 | -0.1973 | No |
| 71 | Grb2 | 8595 | -0.192 | -0.2152 | No |
| 72 | Gnai2 | 8812 | -0.210 | -0.2264 | Yes |
| 73 | Prep | 8818 | -0.211 | -0.2205 | Yes |
| 74 | Prdm4 | 8831 | -0.212 | -0.2152 | Yes |
| 75 | Phex | 8841 | -0.212 | -0.2096 | Yes |
| 76 | Gngt2 | 8901 | -0.216 | -0.2079 | Yes |
| 77 | Psmb9 | 8926 | -0.219 | -0.2033 | Yes |
| 78 | Atox1 | 8987 | -0.224 | -0.2015 | Yes |
| 79 | Dusp5 | 9144 | -0.236 | -0.2071 | Yes |
| 80 | Car2 | 9162 | -0.237 | -0.2014 | Yes |
| 81 | Serpinc1 | 9364 | -0.254 | -0.2101 | Yes |
| 82 | Zeb1 | 9435 | -0.260 | -0.2081 | Yes |
| 83 | Gnb2 | 9523 | -0.266 | -0.2072 | Yes |
| 84 | Ctsl | 9543 | -0.268 | -0.2007 | Yes |
| 85 | Dock10 | 9798 | -0.289 | -0.2127 | Yes |
| 86 | Ppp2cb | 9799 | -0.289 | -0.2041 | Yes |
| 87 | Dpp4 | 9895 | -0.298 | -0.2029 | Yes |
| 88 | Itgam | 10001 | -0.309 | -0.2022 | Yes |
| 89 | Rabif | 10033 | -0.312 | -0.1954 | Yes |
| 90 | Ctso | 10101 | -0.318 | -0.1913 | Yes |
| 91 | Lyn | 10352 | -0.345 | -0.2013 | Yes |
| 92 | Lgals3 | 10520 | -0.363 | -0.2040 | Yes |
| 93 | Stx4a | 10619 | -0.375 | -0.2008 | Yes |
| 94 | Cr2 | 10673 | -0.380 | -0.1937 | Yes |
| 95 | Rhog | 10730 | -0.387 | -0.1867 | Yes |
| 96 | F8 | 10733 | -0.387 | -0.1753 | Yes |
| 97 | Dusp6 | 10836 | -0.400 | -0.1716 | Yes |
| 98 | Plscr1 | 10850 | -0.401 | -0.1607 | Yes |
| 99 | Irf7 | 10878 | -0.406 | -0.1508 | Yes |
| 100 | Cd46 | 10883 | -0.406 | -0.1390 | Yes |
| 101 | Ctsd | 10893 | -0.408 | -0.1275 | Yes |
| 102 | C2 | 11044 | -0.426 | -0.1270 | Yes |
| 103 | Gp1ba | 11203 | -0.448 | -0.1264 | Yes |
| 104 | Csrp1 | 11339 | -0.468 | -0.1234 | Yes |
| 105 | Casp9 | 11342 | -0.469 | -0.1095 | Yes |
| 106 | Src | 11359 | -0.471 | -0.0968 | Yes |
| 107 | Casp4 | 11690 | -0.534 | -0.1076 | Yes |
| 108 | Pfn1 | 11707 | -0.539 | -0.0928 | Yes |
| 109 | Timp2 | 11769 | -0.554 | -0.0812 | Yes |
| 110 | Gpd2 | 11825 | -0.570 | -0.0686 | Yes |
| 111 | Usp14 | 11841 | -0.574 | -0.0527 | Yes |
| 112 | Fyn | 11883 | -0.588 | -0.0384 | Yes |
| 113 | Mmp14 | 12023 | -0.634 | -0.0308 | Yes |
| 114 | Msrb1 | 12057 | -0.648 | -0.0141 | Yes |
| 115 | Gata3 | 12166 | -0.705 | -0.0018 | Yes |
| 116 | Cpq | 12288 | -0.805 | 0.0124 | Yes |