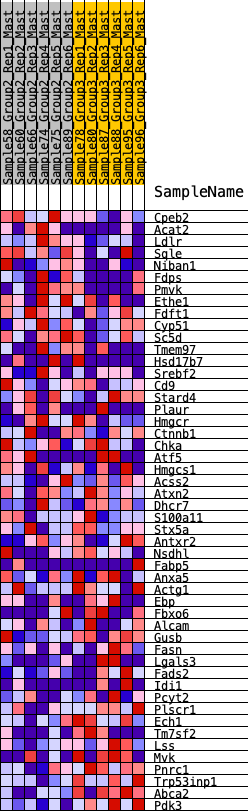
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
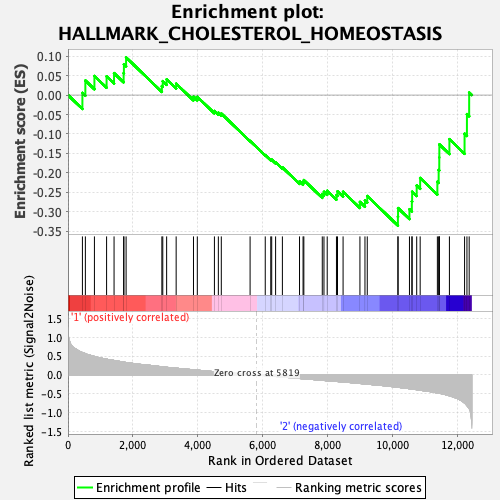
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.33589092 |
| Normalized Enrichment Score (NES) | -1.2597337 |
| Nominal p-value | 0.12153518 |
| FDR q-value | 0.8203252 |
| FWER p-Value | 0.872 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpeb2 | 444 | 0.594 | 0.0055 | No |
| 2 | Acat2 | 535 | 0.562 | 0.0374 | No |
| 3 | Ldlr | 814 | 0.489 | 0.0490 | No |
| 4 | Sqle | 1189 | 0.420 | 0.0481 | No |
| 5 | Niban1 | 1419 | 0.384 | 0.0564 | No |
| 6 | Fdps | 1714 | 0.340 | 0.0563 | No |
| 7 | Pmvk | 1723 | 0.338 | 0.0792 | No |
| 8 | Ethe1 | 1790 | 0.329 | 0.0968 | No |
| 9 | Fdft1 | 2892 | 0.216 | 0.0231 | No |
| 10 | Cyp51 | 2920 | 0.214 | 0.0358 | No |
| 11 | Sc5d | 3039 | 0.203 | 0.0404 | No |
| 12 | Tmem97 | 3332 | 0.179 | 0.0293 | No |
| 13 | Hsd17b7 | 3866 | 0.138 | -0.0041 | No |
| 14 | Srebf2 | 3984 | 0.129 | -0.0045 | No |
| 15 | Cd9 | 4511 | 0.086 | -0.0410 | No |
| 16 | Stard4 | 4632 | 0.077 | -0.0453 | No |
| 17 | Plaur | 4723 | 0.072 | -0.0476 | No |
| 18 | Hmgcr | 5610 | 0.014 | -0.1181 | No |
| 19 | Ctnnb1 | 6077 | -0.017 | -0.1545 | No |
| 20 | Chka | 6247 | -0.029 | -0.1661 | No |
| 21 | Atf5 | 6276 | -0.031 | -0.1662 | No |
| 22 | Hmgcs1 | 6396 | -0.040 | -0.1730 | No |
| 23 | Acss2 | 6605 | -0.054 | -0.1861 | No |
| 24 | Atxn2 | 7133 | -0.093 | -0.2221 | No |
| 25 | Dhcr7 | 7242 | -0.099 | -0.2240 | No |
| 26 | S100a11 | 7266 | -0.100 | -0.2188 | No |
| 27 | Stx5a | 7834 | -0.140 | -0.2548 | No |
| 28 | Antxr2 | 7886 | -0.144 | -0.2489 | No |
| 29 | Nsdhl | 7986 | -0.152 | -0.2463 | No |
| 30 | Fabp5 | 8271 | -0.171 | -0.2573 | No |
| 31 | Anxa5 | 8303 | -0.173 | -0.2478 | No |
| 32 | Actg1 | 8475 | -0.184 | -0.2488 | No |
| 33 | Ebp | 8994 | -0.224 | -0.2750 | No |
| 34 | Fbxo6 | 9149 | -0.236 | -0.2709 | No |
| 35 | Alcam | 9220 | -0.241 | -0.2598 | No |
| 36 | Gusb | 10164 | -0.325 | -0.3133 | Yes |
| 37 | Fasn | 10169 | -0.326 | -0.2909 | Yes |
| 38 | Lgals3 | 10520 | -0.363 | -0.2939 | Yes |
| 39 | Fads2 | 10594 | -0.371 | -0.2739 | Yes |
| 40 | Idi1 | 10603 | -0.373 | -0.2486 | Yes |
| 41 | Pcyt2 | 10743 | -0.388 | -0.2328 | Yes |
| 42 | Plscr1 | 10850 | -0.401 | -0.2134 | Yes |
| 43 | Ech1 | 11380 | -0.474 | -0.2230 | Yes |
| 44 | Tm7sf2 | 11420 | -0.481 | -0.1927 | Yes |
| 45 | Lss | 11439 | -0.483 | -0.1605 | Yes |
| 46 | Mvk | 11441 | -0.483 | -0.1269 | Yes |
| 47 | Pnrc1 | 11751 | -0.550 | -0.1135 | Yes |
| 48 | Trp53inp1 | 12218 | -0.741 | -0.0995 | Yes |
| 49 | Abca2 | 12291 | -0.806 | -0.0492 | Yes |
| 50 | Pdk3 | 12360 | -0.878 | 0.0065 | Yes |