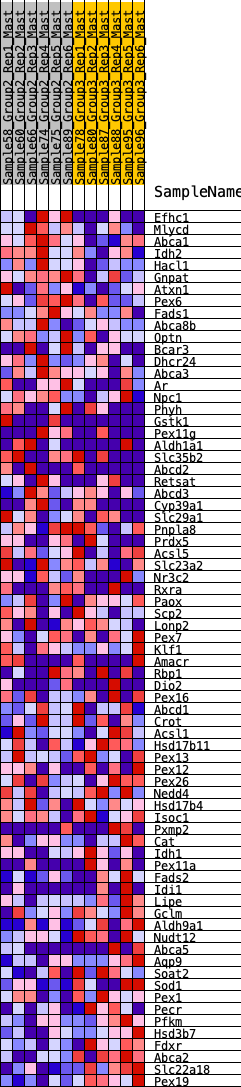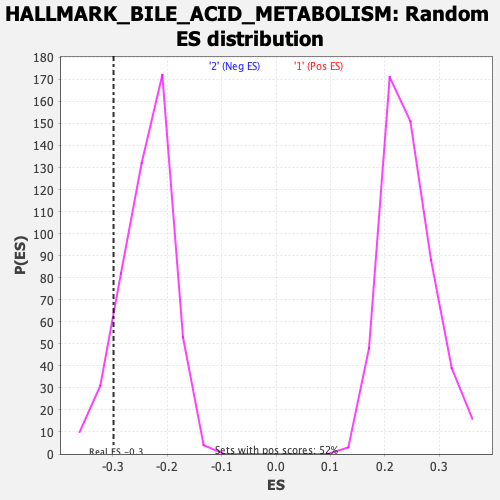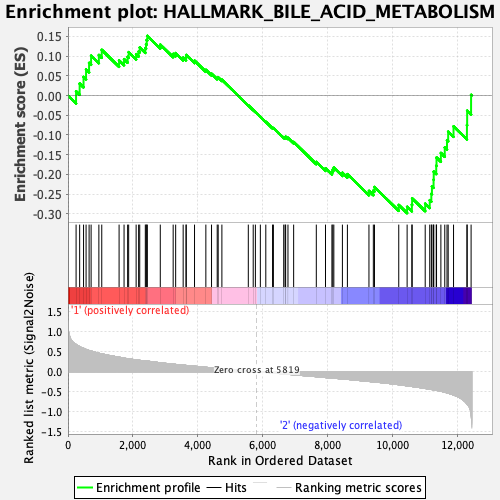
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.29807043 |
| Normalized Enrichment Score (NES) | -1.2571127 |
| Nominal p-value | 0.09090909 |
| FDR q-value | 0.62571865 |
| FWER p-Value | 0.873 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Efhc1 | 249 | 0.681 | 0.0108 | No |
| 2 | Mlycd | 359 | 0.626 | 0.0304 | No |
| 3 | Abca1 | 479 | 0.584 | 0.0472 | No |
| 4 | Idh2 | 557 | 0.556 | 0.0662 | No |
| 5 | Hacl1 | 650 | 0.529 | 0.0828 | No |
| 6 | Gnpat | 712 | 0.515 | 0.1012 | No |
| 7 | Atxn1 | 951 | 0.460 | 0.1028 | No |
| 8 | Pex6 | 1040 | 0.444 | 0.1159 | No |
| 9 | Fads1 | 1574 | 0.360 | 0.0891 | No |
| 10 | Abca8b | 1727 | 0.338 | 0.0922 | No |
| 11 | Optn | 1837 | 0.324 | 0.0981 | No |
| 12 | Bcar3 | 1869 | 0.320 | 0.1100 | No |
| 13 | Dhcr24 | 2099 | 0.294 | 0.1049 | No |
| 14 | Abca3 | 2176 | 0.285 | 0.1117 | No |
| 15 | Ar | 2210 | 0.282 | 0.1218 | No |
| 16 | Npc1 | 2382 | 0.268 | 0.1201 | No |
| 17 | Phyh | 2405 | 0.266 | 0.1304 | No |
| 18 | Gstk1 | 2424 | 0.264 | 0.1409 | No |
| 19 | Pex11g | 2444 | 0.263 | 0.1513 | No |
| 20 | Aldh1a1 | 2843 | 0.222 | 0.1292 | No |
| 21 | Slc35b2 | 3240 | 0.186 | 0.1056 | No |
| 22 | Abcd2 | 3317 | 0.180 | 0.1076 | No |
| 23 | Retsat | 3548 | 0.161 | 0.0964 | No |
| 24 | Abcd3 | 3637 | 0.154 | 0.0962 | No |
| 25 | Cyp39a1 | 3642 | 0.154 | 0.1029 | No |
| 26 | Slc29a1 | 3898 | 0.135 | 0.0884 | No |
| 27 | Pnpla8 | 4247 | 0.108 | 0.0652 | No |
| 28 | Prdx5 | 4422 | 0.093 | 0.0553 | No |
| 29 | Acsl5 | 4597 | 0.079 | 0.0449 | No |
| 30 | Slc23a2 | 4627 | 0.077 | 0.0460 | No |
| 31 | Nr3c2 | 4741 | 0.071 | 0.0401 | No |
| 32 | Rxra | 5555 | 0.017 | -0.0248 | No |
| 33 | Paox | 5706 | 0.007 | -0.0366 | No |
| 34 | Scp2 | 5773 | 0.003 | -0.0419 | No |
| 35 | Lonp2 | 5926 | -0.006 | -0.0539 | No |
| 36 | Pex7 | 6094 | -0.018 | -0.0665 | No |
| 37 | Klf1 | 6302 | -0.032 | -0.0818 | No |
| 38 | Amacr | 6328 | -0.034 | -0.0823 | No |
| 39 | Rbp1 | 6646 | -0.057 | -0.1053 | No |
| 40 | Dio2 | 6688 | -0.060 | -0.1059 | No |
| 41 | Pex16 | 6705 | -0.061 | -0.1044 | No |
| 42 | Abcd1 | 6777 | -0.067 | -0.1072 | No |
| 43 | Crot | 6952 | -0.080 | -0.1176 | No |
| 44 | Acsl1 | 7650 | -0.126 | -0.1682 | No |
| 45 | Hsd17b11 | 7933 | -0.148 | -0.1843 | No |
| 46 | Pex13 | 8131 | -0.161 | -0.1929 | No |
| 47 | Pex12 | 8158 | -0.163 | -0.1876 | No |
| 48 | Pex26 | 8190 | -0.165 | -0.1826 | No |
| 49 | Nedd4 | 8454 | -0.183 | -0.1956 | No |
| 50 | Hsd17b4 | 8608 | -0.193 | -0.1992 | No |
| 51 | Isoc1 | 9272 | -0.246 | -0.2417 | No |
| 52 | Pxmp2 | 9407 | -0.257 | -0.2408 | No |
| 53 | Cat | 9442 | -0.260 | -0.2318 | No |
| 54 | Idh1 | 10190 | -0.328 | -0.2773 | No |
| 55 | Pex11a | 10448 | -0.354 | -0.2820 | Yes |
| 56 | Fads2 | 10594 | -0.371 | -0.2769 | Yes |
| 57 | Idi1 | 10603 | -0.373 | -0.2606 | Yes |
| 58 | Lipe | 11005 | -0.421 | -0.2739 | Yes |
| 59 | Gclm | 11145 | -0.438 | -0.2653 | Yes |
| 60 | Aldh9a1 | 11194 | -0.446 | -0.2489 | Yes |
| 61 | Nudt12 | 11213 | -0.449 | -0.2300 | Yes |
| 62 | Abca5 | 11263 | -0.456 | -0.2133 | Yes |
| 63 | Aqp9 | 11268 | -0.457 | -0.1929 | Yes |
| 64 | Soat2 | 11345 | -0.469 | -0.1778 | Yes |
| 65 | Sod1 | 11354 | -0.471 | -0.1570 | Yes |
| 66 | Pex1 | 11488 | -0.491 | -0.1455 | Yes |
| 67 | Pecr | 11608 | -0.515 | -0.1318 | Yes |
| 68 | Pfkm | 11677 | -0.533 | -0.1131 | Yes |
| 69 | Hsd3b7 | 11713 | -0.541 | -0.0914 | Yes |
| 70 | Fdxr | 11878 | -0.587 | -0.0780 | Yes |
| 71 | Abca2 | 12291 | -0.806 | -0.0748 | Yes |
| 72 | Slc22a18 | 12298 | -0.817 | -0.0382 | Yes |
| 73 | Pex19 | 12419 | -1.095 | 0.0018 | Yes |