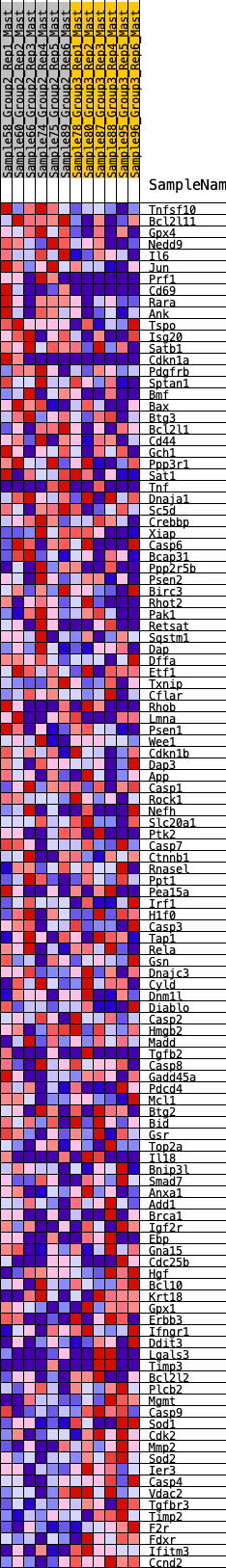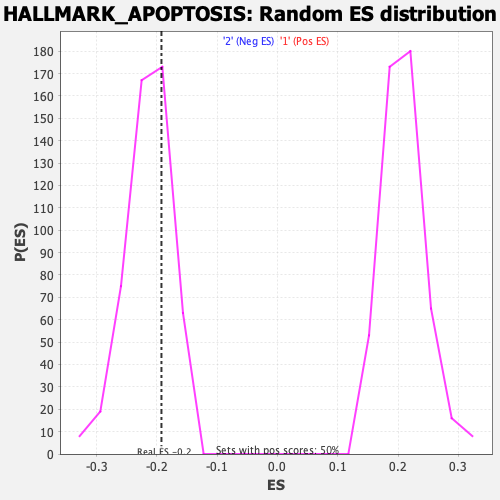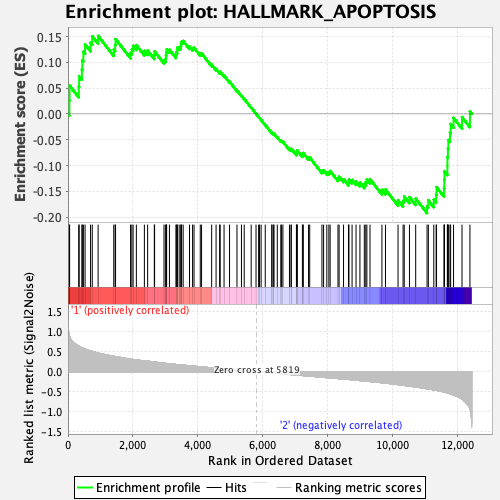
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.19239251 |
| Normalized Enrichment Score (NES) | -0.9018043 |
| Nominal p-value | 0.6792079 |
| FDR q-value | 0.9875123 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfsf10 | 43 | 0.909 | 0.0262 | No |
| 2 | Bcl2l11 | 50 | 0.891 | 0.0549 | No |
| 3 | Gpx4 | 332 | 0.638 | 0.0529 | No |
| 4 | Nedd9 | 343 | 0.633 | 0.0728 | No |
| 5 | Il6 | 428 | 0.598 | 0.0856 | No |
| 6 | Jun | 441 | 0.595 | 0.1041 | No |
| 7 | Prf1 | 474 | 0.585 | 0.1206 | No |
| 8 | Cd69 | 526 | 0.566 | 0.1350 | No |
| 9 | Rara | 695 | 0.518 | 0.1383 | No |
| 10 | Ank | 747 | 0.507 | 0.1507 | No |
| 11 | Tspo | 928 | 0.465 | 0.1513 | No |
| 12 | Isg20 | 1410 | 0.386 | 0.1249 | No |
| 13 | Satb1 | 1452 | 0.379 | 0.1339 | No |
| 14 | Cdkn1a | 1465 | 0.376 | 0.1453 | No |
| 15 | Pdgfrb | 1927 | 0.313 | 0.1181 | No |
| 16 | Sptan1 | 1967 | 0.309 | 0.1250 | No |
| 17 | Bmf | 2006 | 0.304 | 0.1319 | No |
| 18 | Bax | 2106 | 0.294 | 0.1335 | No |
| 19 | Btg3 | 2353 | 0.270 | 0.1223 | No |
| 20 | Bcl2l1 | 2454 | 0.262 | 0.1228 | No |
| 21 | Cd44 | 2659 | 0.239 | 0.1141 | No |
| 22 | Gch1 | 2668 | 0.239 | 0.1212 | No |
| 23 | Ppp3r1 | 2958 | 0.210 | 0.1046 | No |
| 24 | Sat1 | 3013 | 0.206 | 0.1070 | No |
| 25 | Tnf | 3018 | 0.205 | 0.1134 | No |
| 26 | Dnaja1 | 3029 | 0.205 | 0.1192 | No |
| 27 | Sc5d | 3039 | 0.203 | 0.1252 | No |
| 28 | Crebbp | 3125 | 0.196 | 0.1247 | No |
| 29 | Xiap | 3329 | 0.179 | 0.1141 | No |
| 30 | Casp6 | 3336 | 0.178 | 0.1194 | No |
| 31 | Bcap31 | 3366 | 0.177 | 0.1228 | No |
| 32 | Ppp2r5b | 3367 | 0.177 | 0.1286 | No |
| 33 | Psen2 | 3425 | 0.172 | 0.1296 | No |
| 34 | Birc3 | 3476 | 0.167 | 0.1310 | No |
| 35 | Rhot2 | 3483 | 0.167 | 0.1360 | No |
| 36 | Pak1 | 3499 | 0.165 | 0.1402 | No |
| 37 | Retsat | 3548 | 0.161 | 0.1415 | No |
| 38 | Sqstm1 | 3744 | 0.147 | 0.1305 | No |
| 39 | Dap | 3838 | 0.140 | 0.1275 | No |
| 40 | Dffa | 3883 | 0.136 | 0.1284 | No |
| 41 | Etf1 | 4075 | 0.121 | 0.1168 | No |
| 42 | Txnip | 4115 | 0.117 | 0.1175 | No |
| 43 | Cflar | 4426 | 0.093 | 0.0954 | No |
| 44 | Rhob | 4562 | 0.081 | 0.0871 | No |
| 45 | Lmna | 4672 | 0.075 | 0.0807 | No |
| 46 | Psen1 | 4688 | 0.074 | 0.0819 | No |
| 47 | Wee1 | 4808 | 0.067 | 0.0744 | No |
| 48 | Cdkn1b | 4977 | 0.056 | 0.0626 | No |
| 49 | Dap3 | 5206 | 0.039 | 0.0454 | No |
| 50 | App | 5344 | 0.031 | 0.0353 | No |
| 51 | Casp1 | 5429 | 0.025 | 0.0293 | No |
| 52 | Rock1 | 5643 | 0.011 | 0.0124 | No |
| 53 | Nefh | 5793 | 0.002 | 0.0003 | No |
| 54 | Slc20a1 | 5873 | -0.003 | -0.0060 | No |
| 55 | Ptk2 | 5901 | -0.005 | -0.0080 | No |
| 56 | Casp7 | 5950 | -0.008 | -0.0116 | No |
| 57 | Ctnnb1 | 6077 | -0.017 | -0.0213 | No |
| 58 | Rnasel | 6278 | -0.031 | -0.0365 | No |
| 59 | Ppt1 | 6286 | -0.032 | -0.0360 | No |
| 60 | Pea15a | 6339 | -0.035 | -0.0391 | No |
| 61 | Irf1 | 6343 | -0.035 | -0.0382 | No |
| 62 | H1f0 | 6448 | -0.043 | -0.0452 | No |
| 63 | Casp3 | 6556 | -0.051 | -0.0522 | No |
| 64 | Tap1 | 6581 | -0.053 | -0.0525 | No |
| 65 | Rela | 6621 | -0.055 | -0.0538 | No |
| 66 | Gsn | 6823 | -0.071 | -0.0678 | No |
| 67 | Dnajc3 | 6872 | -0.074 | -0.0693 | No |
| 68 | Cyld | 6879 | -0.074 | -0.0674 | No |
| 69 | Dnm1l | 7037 | -0.085 | -0.0773 | No |
| 70 | Diablo | 7041 | -0.086 | -0.0748 | No |
| 71 | Casp2 | 7059 | -0.087 | -0.0733 | No |
| 72 | Hmgb2 | 7061 | -0.088 | -0.0705 | No |
| 73 | Madd | 7221 | -0.098 | -0.0802 | No |
| 74 | Tgfb2 | 7236 | -0.099 | -0.0781 | No |
| 75 | Casp8 | 7247 | -0.099 | -0.0757 | No |
| 76 | Gadd45a | 7411 | -0.111 | -0.0853 | No |
| 77 | Pdcd4 | 7447 | -0.113 | -0.0844 | No |
| 78 | Mcl1 | 7824 | -0.140 | -0.1104 | No |
| 79 | Btg2 | 7875 | -0.144 | -0.1098 | No |
| 80 | Bid | 7974 | -0.151 | -0.1128 | No |
| 81 | Gsr | 8037 | -0.155 | -0.1127 | No |
| 82 | Top2a | 8076 | -0.158 | -0.1106 | No |
| 83 | Il18 | 8318 | -0.174 | -0.1245 | No |
| 84 | Bnip3l | 8352 | -0.176 | -0.1214 | No |
| 85 | Smad7 | 8489 | -0.185 | -0.1264 | No |
| 86 | Anxa1 | 8641 | -0.196 | -0.1323 | No |
| 87 | Add1 | 8656 | -0.198 | -0.1269 | No |
| 88 | Brca1 | 8752 | -0.205 | -0.1279 | No |
| 89 | Igf2r | 8874 | -0.214 | -0.1308 | No |
| 90 | Ebp | 8994 | -0.224 | -0.1331 | No |
| 91 | Gna15 | 9129 | -0.234 | -0.1363 | No |
| 92 | Cdc25b | 9174 | -0.239 | -0.1321 | No |
| 93 | Hgf | 9208 | -0.240 | -0.1269 | No |
| 94 | Bcl10 | 9304 | -0.249 | -0.1265 | No |
| 95 | Krt18 | 9672 | -0.278 | -0.1472 | No |
| 96 | Gpx1 | 9781 | -0.287 | -0.1465 | No |
| 97 | Erbb3 | 10168 | -0.326 | -0.1672 | No |
| 98 | Ifngr1 | 10323 | -0.342 | -0.1685 | No |
| 99 | Ddit3 | 10359 | -0.346 | -0.1600 | No |
| 100 | Lgals3 | 10520 | -0.363 | -0.1612 | No |
| 101 | Timp3 | 10712 | -0.385 | -0.1641 | No |
| 102 | Bcl2l2 | 11062 | -0.428 | -0.1784 | Yes |
| 103 | Plcb2 | 11102 | -0.433 | -0.1674 | Yes |
| 104 | Mgmt | 11270 | -0.457 | -0.1660 | Yes |
| 105 | Casp9 | 11342 | -0.469 | -0.1564 | Yes |
| 106 | Sod1 | 11354 | -0.471 | -0.1419 | Yes |
| 107 | Cdk2 | 11585 | -0.509 | -0.1440 | Yes |
| 108 | Mmp2 | 11593 | -0.512 | -0.1278 | Yes |
| 109 | Sod2 | 11599 | -0.513 | -0.1114 | Yes |
| 110 | Ier3 | 11689 | -0.534 | -0.1012 | Yes |
| 111 | Casp4 | 11690 | -0.534 | -0.0837 | Yes |
| 112 | Vdac2 | 11706 | -0.539 | -0.0673 | Yes |
| 113 | Tgfbr3 | 11719 | -0.543 | -0.0505 | Yes |
| 114 | Timp2 | 11769 | -0.554 | -0.0364 | Yes |
| 115 | F2r | 11789 | -0.560 | -0.0196 | Yes |
| 116 | Fdxr | 11878 | -0.587 | -0.0076 | Yes |
| 117 | Ifitm3 | 12139 | -0.691 | -0.0061 | Yes |
| 118 | Ccnd2 | 12383 | -0.932 | 0.0047 | Yes |