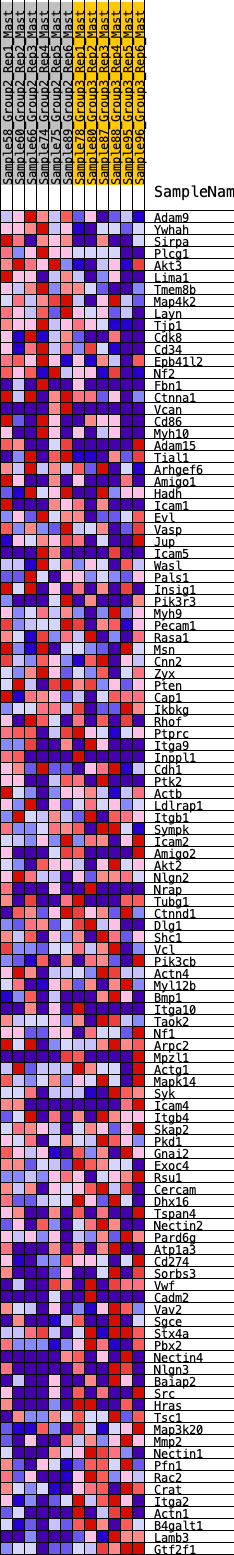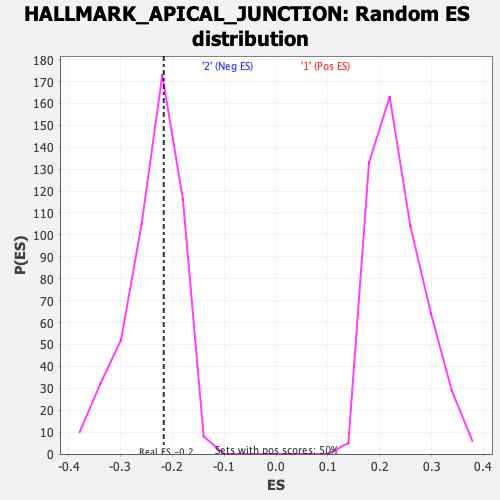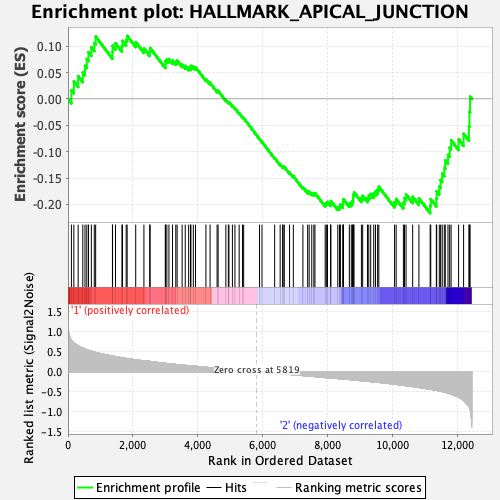
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group3.Mast_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.21646051 |
| Normalized Enrichment Score (NES) | -0.91473883 |
| Nominal p-value | 0.5947581 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adam9 | 105 | 0.786 | 0.0164 | No |
| 2 | Ywhah | 180 | 0.718 | 0.0332 | No |
| 3 | Sirpa | 313 | 0.649 | 0.0431 | No |
| 4 | Plcg1 | 456 | 0.590 | 0.0503 | No |
| 5 | Akt3 | 524 | 0.567 | 0.0629 | No |
| 6 | Lima1 | 580 | 0.547 | 0.0757 | No |
| 7 | Tmem8b | 631 | 0.534 | 0.0886 | No |
| 8 | Map4k2 | 719 | 0.512 | 0.0978 | No |
| 9 | Layn | 810 | 0.492 | 0.1061 | No |
| 10 | Tjp1 | 852 | 0.480 | 0.1180 | No |
| 11 | Cdk8 | 1369 | 0.391 | 0.0886 | No |
| 12 | Cd34 | 1373 | 0.391 | 0.1007 | No |
| 13 | Epb41l2 | 1463 | 0.377 | 0.1055 | No |
| 14 | Nf2 | 1662 | 0.348 | 0.1005 | No |
| 15 | Fbn1 | 1680 | 0.345 | 0.1100 | No |
| 16 | Ctnna1 | 1792 | 0.329 | 0.1115 | No |
| 17 | Vcan | 1822 | 0.325 | 0.1194 | No |
| 18 | Cd86 | 2086 | 0.295 | 0.1075 | No |
| 19 | Myh10 | 2339 | 0.271 | 0.0956 | No |
| 20 | Adam15 | 2517 | 0.256 | 0.0894 | No |
| 21 | Tial1 | 2530 | 0.254 | 0.0965 | No |
| 22 | Arhgef6 | 2999 | 0.207 | 0.0651 | No |
| 23 | Amigo1 | 3002 | 0.206 | 0.0715 | No |
| 24 | Hadh | 3045 | 0.203 | 0.0745 | No |
| 25 | Icam1 | 3110 | 0.197 | 0.0756 | No |
| 26 | Evl | 3220 | 0.188 | 0.0727 | No |
| 27 | Vasp | 3321 | 0.180 | 0.0703 | No |
| 28 | Jup | 3362 | 0.177 | 0.0727 | No |
| 29 | Icam5 | 3516 | 0.164 | 0.0654 | No |
| 30 | Wasl | 3612 | 0.157 | 0.0627 | No |
| 31 | Pals1 | 3717 | 0.149 | 0.0590 | No |
| 32 | Insig1 | 3773 | 0.144 | 0.0591 | No |
| 33 | Pik3r3 | 3787 | 0.143 | 0.0626 | No |
| 34 | Myh9 | 3861 | 0.138 | 0.0611 | No |
| 35 | Pecam1 | 3929 | 0.132 | 0.0599 | No |
| 36 | Rasa1 | 4249 | 0.108 | 0.0374 | No |
| 37 | Msn | 4377 | 0.097 | 0.0302 | No |
| 38 | Cnn2 | 4593 | 0.079 | 0.0153 | No |
| 39 | Zyx | 4625 | 0.077 | 0.0152 | No |
| 40 | Pten | 4863 | 0.063 | -0.0020 | No |
| 41 | Cap1 | 4940 | 0.059 | -0.0063 | No |
| 42 | Ikbkg | 4958 | 0.057 | -0.0059 | No |
| 43 | Rhof | 5065 | 0.050 | -0.0129 | No |
| 44 | Ptprc | 5143 | 0.044 | -0.0177 | No |
| 45 | Itga9 | 5273 | 0.035 | -0.0270 | No |
| 46 | Inppl1 | 5376 | 0.029 | -0.0344 | No |
| 47 | Cdh1 | 5412 | 0.027 | -0.0364 | No |
| 48 | Ptk2 | 5901 | -0.005 | -0.0758 | No |
| 49 | Actb | 5977 | -0.011 | -0.0816 | No |
| 50 | Ldlrap1 | 6367 | -0.037 | -0.1119 | No |
| 51 | Itgb1 | 6536 | -0.050 | -0.1240 | No |
| 52 | Sympk | 6611 | -0.054 | -0.1283 | No |
| 53 | Icam2 | 6627 | -0.055 | -0.1277 | No |
| 54 | Amigo2 | 6664 | -0.058 | -0.1288 | No |
| 55 | Akt2 | 6826 | -0.071 | -0.1396 | No |
| 56 | Nlgn2 | 6942 | -0.079 | -0.1464 | No |
| 57 | Nrap | 7237 | -0.099 | -0.1671 | No |
| 58 | Tubg1 | 7384 | -0.109 | -0.1755 | No |
| 59 | Ctnnd1 | 7430 | -0.112 | -0.1756 | No |
| 60 | Dlg1 | 7510 | -0.117 | -0.1783 | No |
| 61 | Shc1 | 7575 | -0.121 | -0.1796 | No |
| 62 | Vcl | 7608 | -0.124 | -0.1783 | No |
| 63 | Pik3cb | 7924 | -0.148 | -0.1992 | No |
| 64 | Actn4 | 7959 | -0.150 | -0.1972 | No |
| 65 | Myl12b | 7995 | -0.153 | -0.1952 | No |
| 66 | Bmp1 | 8090 | -0.159 | -0.1978 | No |
| 67 | Itga10 | 8099 | -0.159 | -0.1934 | No |
| 68 | Taok2 | 8312 | -0.173 | -0.2051 | Yes |
| 69 | Nf1 | 8364 | -0.177 | -0.2036 | Yes |
| 70 | Arpc2 | 8393 | -0.179 | -0.2002 | Yes |
| 71 | Mpzl1 | 8468 | -0.184 | -0.2004 | Yes |
| 72 | Actg1 | 8475 | -0.184 | -0.1950 | Yes |
| 73 | Mapk14 | 8487 | -0.185 | -0.1901 | Yes |
| 74 | Syk | 8668 | -0.198 | -0.1984 | Yes |
| 75 | Icam4 | 8728 | -0.203 | -0.1967 | Yes |
| 76 | Itgb4 | 8764 | -0.206 | -0.1930 | Yes |
| 77 | Skap2 | 8786 | -0.209 | -0.1881 | Yes |
| 78 | Pkd1 | 8789 | -0.209 | -0.1816 | Yes |
| 79 | Gnai2 | 8812 | -0.210 | -0.1767 | Yes |
| 80 | Exoc4 | 9040 | -0.228 | -0.1879 | Yes |
| 81 | Rsu1 | 9079 | -0.231 | -0.1837 | Yes |
| 82 | Cercam | 9226 | -0.241 | -0.1879 | Yes |
| 83 | Dhx16 | 9267 | -0.246 | -0.1833 | Yes |
| 84 | Tspan4 | 9326 | -0.251 | -0.1801 | Yes |
| 85 | Nectin2 | 9419 | -0.258 | -0.1793 | Yes |
| 86 | Pard6g | 9473 | -0.262 | -0.1753 | Yes |
| 87 | Atp1a3 | 9539 | -0.267 | -0.1721 | Yes |
| 88 | Cd274 | 9576 | -0.270 | -0.1665 | Yes |
| 89 | Sorbs3 | 10061 | -0.315 | -0.1957 | Yes |
| 90 | Vwf | 10110 | -0.319 | -0.1895 | Yes |
| 91 | Cadm2 | 10333 | -0.343 | -0.1966 | Yes |
| 92 | Vav2 | 10367 | -0.346 | -0.1883 | Yes |
| 93 | Sgce | 10415 | -0.352 | -0.1809 | Yes |
| 94 | Stx4a | 10619 | -0.375 | -0.1855 | Yes |
| 95 | Pbx2 | 10811 | -0.397 | -0.1884 | Yes |
| 96 | Nectin4 | 11158 | -0.440 | -0.2025 | Yes |
| 97 | Nlgn3 | 11176 | -0.442 | -0.1899 | Yes |
| 98 | Baiap2 | 11348 | -0.470 | -0.1888 | Yes |
| 99 | Src | 11359 | -0.471 | -0.1747 | Yes |
| 100 | Hras | 11440 | -0.483 | -0.1659 | Yes |
| 101 | Tsc1 | 11480 | -0.490 | -0.1535 | Yes |
| 102 | Map3k20 | 11529 | -0.499 | -0.1415 | Yes |
| 103 | Mmp2 | 11593 | -0.512 | -0.1304 | Yes |
| 104 | Nectin1 | 11623 | -0.518 | -0.1163 | Yes |
| 105 | Pfn1 | 11707 | -0.539 | -0.1059 | Yes |
| 106 | Rac2 | 11759 | -0.552 | -0.0926 | Yes |
| 107 | Crat | 11803 | -0.563 | -0.0782 | Yes |
| 108 | Itga2 | 12034 | -0.638 | -0.0766 | Yes |
| 109 | Actn1 | 12188 | -0.723 | -0.0661 | Yes |
| 110 | B4galt1 | 12352 | -0.873 | -0.0516 | Yes |
| 111 | Lamb3 | 12368 | -0.904 | -0.0241 | Yes |
| 112 | Gtf2f1 | 12388 | -0.944 | 0.0043 | Yes |