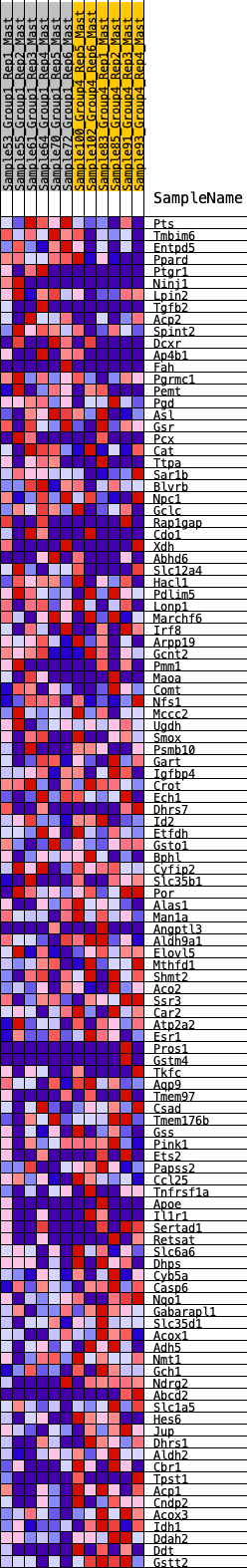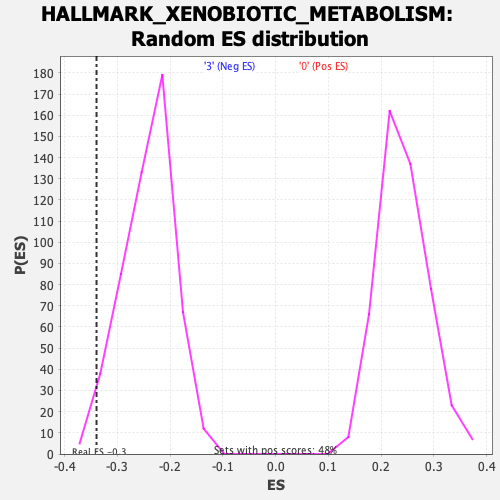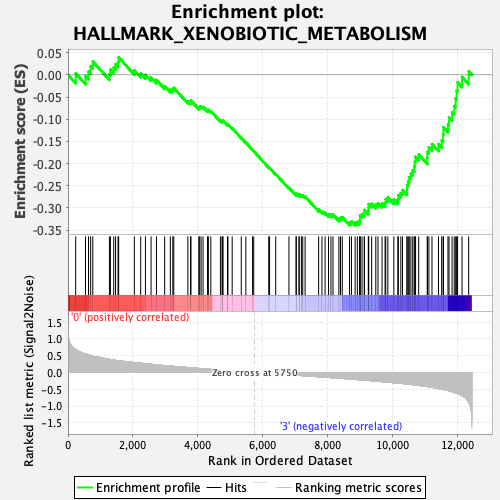
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.3397182 |
| Normalized Enrichment Score (NES) | -1.4071664 |
| Nominal p-value | 0.011560693 |
| FDR q-value | 0.15796098 |
| FWER p-Value | 0.525 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pts | 237 | 0.674 | 0.0032 | No |
| 2 | Tmbim6 | 539 | 0.553 | -0.0027 | No |
| 3 | Entpd5 | 632 | 0.525 | 0.0073 | No |
| 4 | Ppard | 698 | 0.507 | 0.0189 | No |
| 5 | Ptgr1 | 764 | 0.491 | 0.0300 | No |
| 6 | Ninj1 | 1275 | 0.389 | 0.0016 | No |
| 7 | Lpin2 | 1307 | 0.384 | 0.0119 | No |
| 8 | Tgfb2 | 1409 | 0.370 | 0.0160 | No |
| 9 | Acp2 | 1465 | 0.362 | 0.0236 | No |
| 10 | Spint2 | 1542 | 0.351 | 0.0291 | No |
| 11 | Dcxr | 1561 | 0.348 | 0.0393 | No |
| 12 | Ap4b1 | 2044 | 0.294 | 0.0100 | No |
| 13 | Fah | 2240 | 0.274 | 0.0033 | No |
| 14 | Pgrmc1 | 2386 | 0.259 | 0.0002 | No |
| 15 | Pemt | 2559 | 0.243 | -0.0057 | No |
| 16 | Pgd | 2725 | 0.228 | -0.0115 | No |
| 17 | Asl | 2978 | 0.201 | -0.0252 | No |
| 18 | Gsr | 3153 | 0.187 | -0.0331 | No |
| 19 | Pcx | 3229 | 0.183 | -0.0331 | No |
| 20 | Cat | 3259 | 0.180 | -0.0294 | No |
| 21 | Ttpa | 3693 | 0.146 | -0.0597 | No |
| 22 | Sar1b | 3776 | 0.140 | -0.0617 | No |
| 23 | Blvrb | 3788 | 0.139 | -0.0579 | No |
| 24 | Npc1 | 4029 | 0.120 | -0.0734 | No |
| 25 | Gclc | 4056 | 0.117 | -0.0716 | No |
| 26 | Rap1gap | 4100 | 0.115 | -0.0713 | No |
| 27 | Cdo1 | 4161 | 0.112 | -0.0724 | No |
| 28 | Xdh | 4298 | 0.102 | -0.0800 | No |
| 29 | Abhd6 | 4325 | 0.099 | -0.0789 | No |
| 30 | Slc12a4 | 4400 | 0.093 | -0.0818 | No |
| 31 | Hacl1 | 4696 | 0.071 | -0.1033 | No |
| 32 | Pdlim5 | 4745 | 0.067 | -0.1050 | No |
| 33 | Lonp1 | 4768 | 0.066 | -0.1045 | No |
| 34 | Marchf6 | 4773 | 0.066 | -0.1027 | No |
| 35 | Irf8 | 4914 | 0.056 | -0.1122 | No |
| 36 | Arpp19 | 4928 | 0.055 | -0.1114 | No |
| 37 | Gcnt2 | 5058 | 0.048 | -0.1203 | No |
| 38 | Pmm1 | 5338 | 0.027 | -0.1420 | No |
| 39 | Maoa | 5480 | 0.019 | -0.1528 | No |
| 40 | Comt | 5686 | 0.004 | -0.1693 | No |
| 41 | Nfs1 | 5722 | 0.001 | -0.1721 | No |
| 42 | Mccc2 | 6184 | -0.025 | -0.2086 | No |
| 43 | Ugdh | 6211 | -0.027 | -0.2098 | No |
| 44 | Smox | 6399 | -0.040 | -0.2237 | No |
| 45 | Psmb10 | 6807 | -0.065 | -0.2545 | No |
| 46 | Gart | 7026 | -0.078 | -0.2696 | No |
| 47 | Igfbp4 | 7034 | -0.079 | -0.2675 | No |
| 48 | Crot | 7119 | -0.084 | -0.2715 | No |
| 49 | Ech1 | 7120 | -0.084 | -0.2687 | No |
| 50 | Dhrs7 | 7191 | -0.090 | -0.2714 | No |
| 51 | Id2 | 7232 | -0.093 | -0.2716 | No |
| 52 | Etfdh | 7305 | -0.097 | -0.2742 | No |
| 53 | Gsto1 | 7722 | -0.126 | -0.3037 | No |
| 54 | Bphl | 7824 | -0.132 | -0.3075 | No |
| 55 | Cyfip2 | 7920 | -0.140 | -0.3106 | No |
| 56 | Slc35b1 | 8026 | -0.147 | -0.3142 | No |
| 57 | Por | 8101 | -0.152 | -0.3151 | No |
| 58 | Alas1 | 8164 | -0.156 | -0.3150 | No |
| 59 | Man1a | 8346 | -0.168 | -0.3241 | No |
| 60 | Angptl3 | 8396 | -0.172 | -0.3223 | No |
| 61 | Aldh9a1 | 8450 | -0.176 | -0.3208 | No |
| 62 | Elovl5 | 8675 | -0.192 | -0.3325 | No |
| 63 | Mthfd1 | 8733 | -0.196 | -0.3306 | No |
| 64 | Shmt2 | 8846 | -0.206 | -0.3329 | Yes |
| 65 | Aco2 | 8915 | -0.211 | -0.3313 | Yes |
| 66 | Ssr3 | 8982 | -0.215 | -0.3295 | Yes |
| 67 | Car2 | 8999 | -0.216 | -0.3236 | Yes |
| 68 | Atp2a2 | 9002 | -0.216 | -0.3166 | Yes |
| 69 | Esr1 | 9059 | -0.220 | -0.3138 | Yes |
| 70 | Pros1 | 9129 | -0.226 | -0.3118 | Yes |
| 71 | Gstm4 | 9131 | -0.226 | -0.3044 | Yes |
| 72 | Tkfc | 9252 | -0.234 | -0.3063 | Yes |
| 73 | Aqp9 | 9260 | -0.235 | -0.2990 | Yes |
| 74 | Tmem97 | 9263 | -0.235 | -0.2913 | Yes |
| 75 | Csad | 9356 | -0.244 | -0.2907 | Yes |
| 76 | Tmem176b | 9483 | -0.255 | -0.2924 | Yes |
| 77 | Gss | 9551 | -0.261 | -0.2891 | Yes |
| 78 | Pink1 | 9675 | -0.271 | -0.2901 | Yes |
| 79 | Ets2 | 9769 | -0.279 | -0.2883 | Yes |
| 80 | Papss2 | 9789 | -0.281 | -0.2805 | Yes |
| 81 | Ccl25 | 9854 | -0.287 | -0.2761 | Yes |
| 82 | Tnfrsf1a | 10039 | -0.304 | -0.2809 | Yes |
| 83 | Apoe | 10162 | -0.313 | -0.2804 | Yes |
| 84 | Il1r1 | 10178 | -0.315 | -0.2711 | Yes |
| 85 | Sertad1 | 10254 | -0.322 | -0.2665 | Yes |
| 86 | Retsat | 10305 | -0.326 | -0.2597 | Yes |
| 87 | Slc6a6 | 10444 | -0.341 | -0.2595 | Yes |
| 88 | Dhps | 10445 | -0.341 | -0.2481 | Yes |
| 89 | Cyb5a | 10484 | -0.345 | -0.2397 | Yes |
| 90 | Casp6 | 10515 | -0.349 | -0.2305 | Yes |
| 91 | Nqo1 | 10565 | -0.355 | -0.2226 | Yes |
| 92 | Gabarapl1 | 10621 | -0.363 | -0.2150 | Yes |
| 93 | Slc35d1 | 10667 | -0.368 | -0.2064 | Yes |
| 94 | Acox1 | 10685 | -0.370 | -0.1954 | Yes |
| 95 | Adh5 | 10708 | -0.373 | -0.1848 | Yes |
| 96 | Nmt1 | 10804 | -0.385 | -0.1797 | Yes |
| 97 | Gch1 | 11069 | -0.419 | -0.1872 | Yes |
| 98 | Ndrg2 | 11073 | -0.419 | -0.1735 | Yes |
| 99 | Abcd2 | 11119 | -0.425 | -0.1629 | Yes |
| 100 | Slc1a5 | 11214 | -0.440 | -0.1559 | Yes |
| 101 | Hes6 | 11414 | -0.477 | -0.1562 | Yes |
| 102 | Jup | 11512 | -0.494 | -0.1476 | Yes |
| 103 | Dhrs1 | 11558 | -0.505 | -0.1344 | Yes |
| 104 | Aldh2 | 11567 | -0.506 | -0.1182 | Yes |
| 105 | Cbr1 | 11707 | -0.535 | -0.1116 | Yes |
| 106 | Tpst1 | 11739 | -0.542 | -0.0961 | Yes |
| 107 | Acp1 | 11833 | -0.571 | -0.0846 | Yes |
| 108 | Cndp2 | 11899 | -0.591 | -0.0702 | Yes |
| 109 | Acox3 | 11944 | -0.607 | -0.0535 | Yes |
| 110 | Idh1 | 11973 | -0.617 | -0.0352 | Yes |
| 111 | Ddah2 | 12003 | -0.626 | -0.0167 | Yes |
| 112 | Ddt | 12140 | -0.691 | -0.0047 | Yes |
| 113 | Gstt2 | 12344 | -0.872 | 0.0079 | Yes |