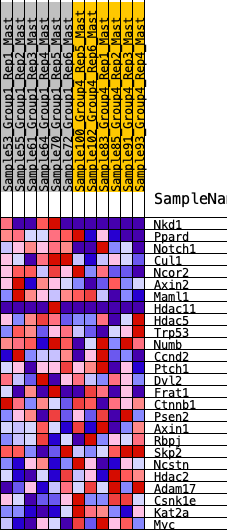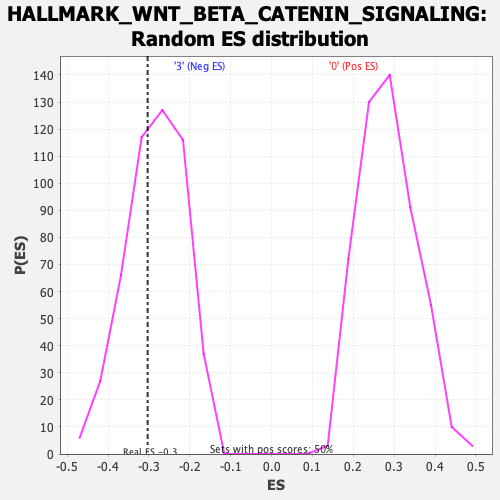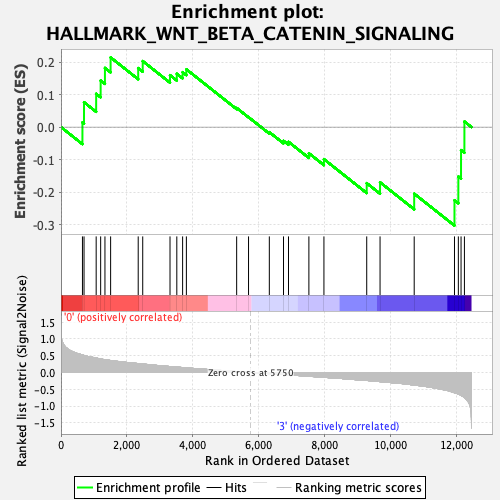
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.30339366 |
| Normalized Enrichment Score (NES) | -1.0752723 |
| Nominal p-value | 0.35282257 |
| FDR q-value | 0.55019575 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nkd1 | 650 | 0.518 | 0.0147 | No |
| 2 | Ppard | 698 | 0.507 | 0.0766 | No |
| 3 | Notch1 | 1066 | 0.430 | 0.1027 | No |
| 4 | Cul1 | 1202 | 0.399 | 0.1436 | No |
| 5 | Ncor2 | 1332 | 0.380 | 0.1824 | No |
| 6 | Axin2 | 1504 | 0.356 | 0.2147 | No |
| 7 | Maml1 | 2340 | 0.265 | 0.1818 | No |
| 8 | Hdac11 | 2477 | 0.251 | 0.2034 | No |
| 9 | Hdac5 | 3304 | 0.178 | 0.1600 | No |
| 10 | Trp53 | 3510 | 0.161 | 0.1644 | No |
| 11 | Numb | 3686 | 0.147 | 0.1692 | No |
| 12 | Ccnd2 | 3800 | 0.138 | 0.1780 | No |
| 13 | Ptch1 | 5324 | 0.028 | 0.0590 | No |
| 14 | Dvl2 | 5684 | 0.004 | 0.0306 | No |
| 15 | Frat1 | 6314 | -0.035 | -0.0156 | No |
| 16 | Ctnnb1 | 6744 | -0.060 | -0.0423 | No |
| 17 | Psen2 | 6895 | -0.070 | -0.0453 | No |
| 18 | Axin1 | 7513 | -0.112 | -0.0806 | No |
| 19 | Rbpj | 7969 | -0.142 | -0.0988 | No |
| 20 | Skp2 | 9266 | -0.235 | -0.1727 | No |
| 21 | Ncstn | 9670 | -0.271 | -0.1701 | No |
| 22 | Hdac2 | 10705 | -0.372 | -0.2051 | No |
| 23 | Adam17 | 11926 | -0.602 | -0.2254 | Yes |
| 24 | Csnk1e | 12043 | -0.637 | -0.1523 | Yes |
| 25 | Kat2a | 12125 | -0.679 | -0.0709 | Yes |
| 26 | Myc | 12227 | -0.743 | 0.0172 | Yes |