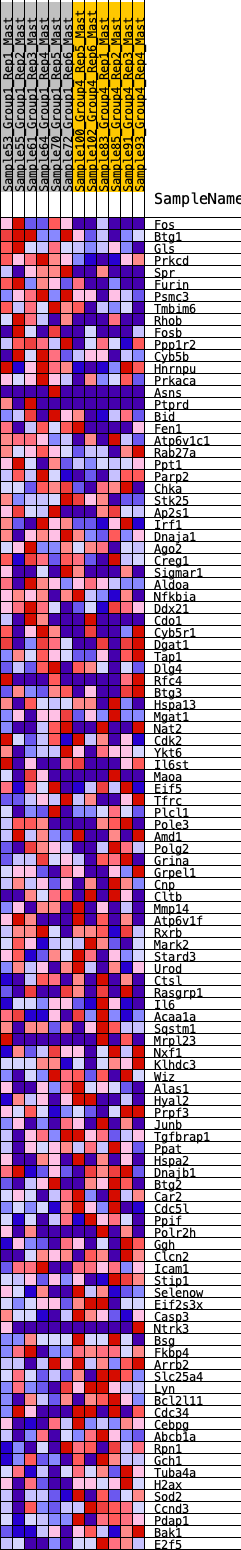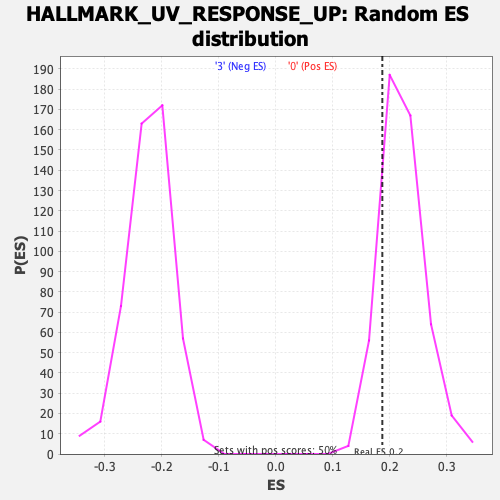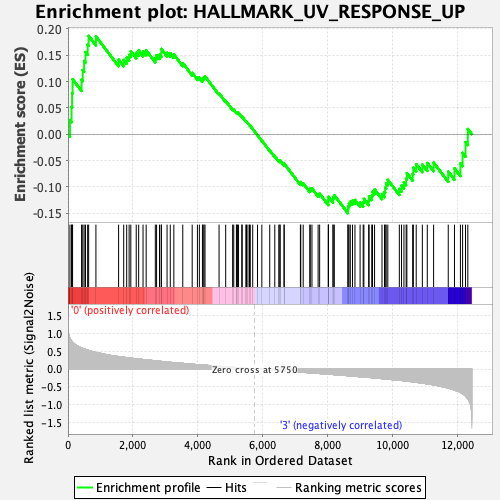
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.18672529 |
| Normalized Enrichment Score (NES) | 0.8361036 |
| Nominal p-value | 0.82902586 |
| FDR q-value | 0.84304863 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fos | 60 | 0.871 | 0.0268 | Yes |
| 2 | Btg1 | 109 | 0.790 | 0.0517 | Yes |
| 3 | Gls | 127 | 0.766 | 0.0782 | Yes |
| 4 | Prkcd | 141 | 0.752 | 0.1045 | Yes |
| 5 | Spr | 418 | 0.594 | 0.1037 | Yes |
| 6 | Furin | 455 | 0.580 | 0.1219 | Yes |
| 7 | Psmc3 | 497 | 0.565 | 0.1391 | Yes |
| 8 | Tmbim6 | 539 | 0.553 | 0.1559 | Yes |
| 9 | Rhob | 605 | 0.535 | 0.1701 | Yes |
| 10 | Fosb | 636 | 0.523 | 0.1867 | Yes |
| 11 | Ppp1r2 | 860 | 0.468 | 0.1857 | No |
| 12 | Cyb5b | 1559 | 0.348 | 0.1417 | No |
| 13 | Hnrnpu | 1716 | 0.330 | 0.1411 | No |
| 14 | Prkaca | 1808 | 0.320 | 0.1454 | No |
| 15 | Asns | 1883 | 0.311 | 0.1507 | No |
| 16 | Ptprd | 1935 | 0.305 | 0.1576 | No |
| 17 | Bid | 2102 | 0.287 | 0.1546 | No |
| 18 | Fen1 | 2177 | 0.280 | 0.1588 | No |
| 19 | Atp6v1c1 | 2312 | 0.268 | 0.1577 | No |
| 20 | Rab27a | 2408 | 0.256 | 0.1593 | No |
| 21 | Ppt1 | 2690 | 0.232 | 0.1449 | No |
| 22 | Parp2 | 2729 | 0.227 | 0.1501 | No |
| 23 | Chka | 2819 | 0.219 | 0.1508 | No |
| 24 | Stk25 | 2874 | 0.213 | 0.1542 | No |
| 25 | Ap2s1 | 2875 | 0.213 | 0.1620 | No |
| 26 | Irf1 | 3052 | 0.196 | 0.1548 | No |
| 27 | Dnaja1 | 3151 | 0.188 | 0.1537 | No |
| 28 | Ago2 | 3261 | 0.180 | 0.1514 | No |
| 29 | Creg1 | 3536 | 0.159 | 0.1350 | No |
| 30 | Sigmar1 | 3826 | 0.136 | 0.1165 | No |
| 31 | Aldoa | 3987 | 0.122 | 0.1080 | No |
| 32 | Nfkbia | 4047 | 0.118 | 0.1075 | No |
| 33 | Ddx21 | 4142 | 0.114 | 0.1040 | No |
| 34 | Cdo1 | 4161 | 0.112 | 0.1066 | No |
| 35 | Cyb5r1 | 4189 | 0.110 | 0.1084 | No |
| 36 | Dgat1 | 4221 | 0.108 | 0.1098 | No |
| 37 | Tap1 | 4654 | 0.075 | 0.0775 | No |
| 38 | Dlg4 | 4858 | 0.060 | 0.0632 | No |
| 39 | Rfc4 | 5072 | 0.047 | 0.0477 | No |
| 40 | Btg3 | 5106 | 0.045 | 0.0466 | No |
| 41 | Hspa13 | 5188 | 0.038 | 0.0414 | No |
| 42 | Mgat1 | 5200 | 0.037 | 0.0419 | No |
| 43 | Nat2 | 5224 | 0.036 | 0.0413 | No |
| 44 | Cdk2 | 5254 | 0.034 | 0.0402 | No |
| 45 | Ykt6 | 5355 | 0.026 | 0.0330 | No |
| 46 | Il6st | 5366 | 0.026 | 0.0331 | No |
| 47 | Maoa | 5480 | 0.019 | 0.0246 | No |
| 48 | Eif5 | 5498 | 0.017 | 0.0239 | No |
| 49 | Tfrc | 5544 | 0.014 | 0.0207 | No |
| 50 | Plcl1 | 5592 | 0.011 | 0.0173 | No |
| 51 | Pole3 | 5616 | 0.009 | 0.0158 | No |
| 52 | Amd1 | 5691 | 0.003 | 0.0099 | No |
| 53 | Polg2 | 5838 | -0.004 | -0.0018 | No |
| 54 | Grina | 5972 | -0.013 | -0.0121 | No |
| 55 | Grpel1 | 6215 | -0.027 | -0.0307 | No |
| 56 | Cnp | 6371 | -0.038 | -0.0419 | No |
| 57 | Cltb | 6494 | -0.046 | -0.0502 | No |
| 58 | Mmp14 | 6528 | -0.048 | -0.0511 | No |
| 59 | Atp6v1f | 6535 | -0.049 | -0.0498 | No |
| 60 | Rxrb | 6657 | -0.055 | -0.0576 | No |
| 61 | Mark2 | 6660 | -0.055 | -0.0558 | No |
| 62 | Stard3 | 7160 | -0.087 | -0.0931 | No |
| 63 | Urod | 7171 | -0.088 | -0.0907 | No |
| 64 | Ctsl | 7246 | -0.094 | -0.0932 | No |
| 65 | Rasgrp1 | 7450 | -0.107 | -0.1058 | No |
| 66 | Il6 | 7463 | -0.108 | -0.1028 | No |
| 67 | Acaa1a | 7516 | -0.112 | -0.1030 | No |
| 68 | Sqstm1 | 7706 | -0.125 | -0.1138 | No |
| 69 | Mrpl23 | 7753 | -0.128 | -0.1129 | No |
| 70 | Nxf1 | 8021 | -0.146 | -0.1292 | No |
| 71 | Klhdc3 | 8022 | -0.146 | -0.1239 | No |
| 72 | Wiz | 8024 | -0.146 | -0.1186 | No |
| 73 | Alas1 | 8164 | -0.156 | -0.1242 | No |
| 74 | Hyal2 | 8168 | -0.156 | -0.1188 | No |
| 75 | Prpf3 | 8205 | -0.159 | -0.1159 | No |
| 76 | Junb | 8623 | -0.188 | -0.1429 | No |
| 77 | Tgfbrap1 | 8630 | -0.188 | -0.1366 | No |
| 78 | Ppat | 8657 | -0.190 | -0.1317 | No |
| 79 | Hspa2 | 8697 | -0.193 | -0.1279 | No |
| 80 | Dnajb1 | 8765 | -0.199 | -0.1261 | No |
| 81 | Btg2 | 8841 | -0.206 | -0.1247 | No |
| 82 | Car2 | 8999 | -0.216 | -0.1295 | No |
| 83 | Cdc5l | 9094 | -0.223 | -0.1291 | No |
| 84 | Ppif | 9116 | -0.225 | -0.1226 | No |
| 85 | Polr2h | 9264 | -0.235 | -0.1260 | No |
| 86 | Ggh | 9275 | -0.236 | -0.1182 | No |
| 87 | Clcn2 | 9359 | -0.244 | -0.1160 | No |
| 88 | Icam1 | 9381 | -0.246 | -0.1088 | No |
| 89 | Stip1 | 9445 | -0.252 | -0.1047 | No |
| 90 | Selenow | 9674 | -0.271 | -0.1134 | No |
| 91 | Eif2s3x | 9751 | -0.278 | -0.1094 | No |
| 92 | Casp3 | 9773 | -0.279 | -0.1010 | No |
| 93 | Ntrk3 | 9803 | -0.283 | -0.0931 | No |
| 94 | Bsg | 9853 | -0.287 | -0.0866 | No |
| 95 | Fkbp4 | 10208 | -0.318 | -0.1037 | No |
| 96 | Arrb2 | 10275 | -0.323 | -0.0973 | No |
| 97 | Slc25a4 | 10350 | -0.331 | -0.0913 | No |
| 98 | Lyn | 10418 | -0.338 | -0.0844 | No |
| 99 | Bcl2l11 | 10440 | -0.341 | -0.0737 | No |
| 100 | Cdc34 | 10619 | -0.363 | -0.0750 | No |
| 101 | Cebpg | 10640 | -0.365 | -0.0633 | No |
| 102 | Abcb1a | 10728 | -0.376 | -0.0567 | No |
| 103 | Rpn1 | 10917 | -0.398 | -0.0574 | No |
| 104 | Gch1 | 11069 | -0.419 | -0.0545 | No |
| 105 | Tuba4a | 11264 | -0.449 | -0.0539 | No |
| 106 | H2ax | 11715 | -0.536 | -0.0708 | No |
| 107 | Sod2 | 11907 | -0.595 | -0.0647 | No |
| 108 | Ccnd3 | 12090 | -0.661 | -0.0554 | No |
| 109 | Pdap1 | 12157 | -0.698 | -0.0354 | No |
| 110 | Bak1 | 12247 | -0.760 | -0.0149 | No |
| 111 | E2f5 | 12318 | -0.841 | 0.0100 | No |