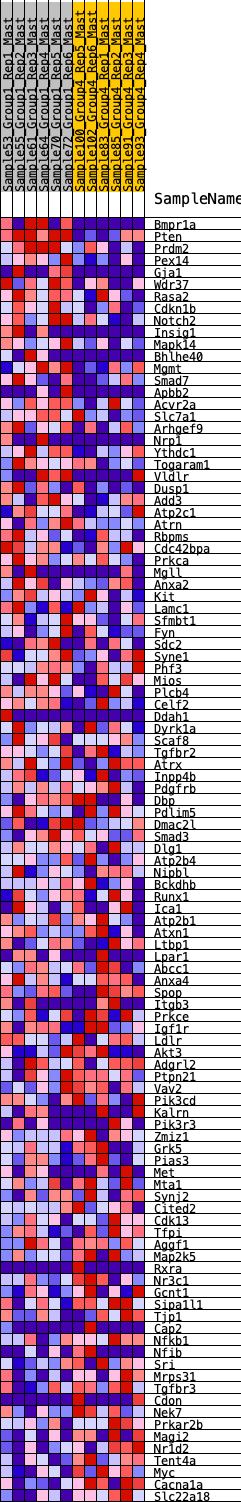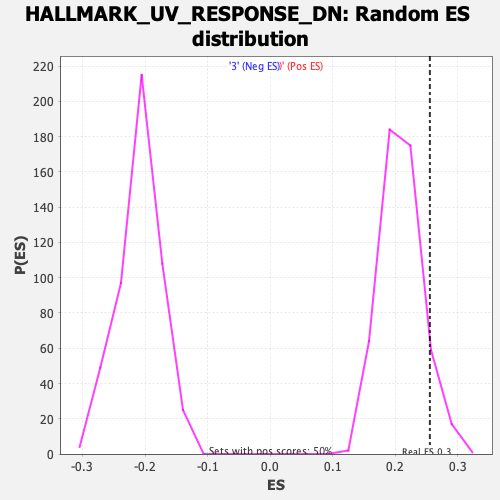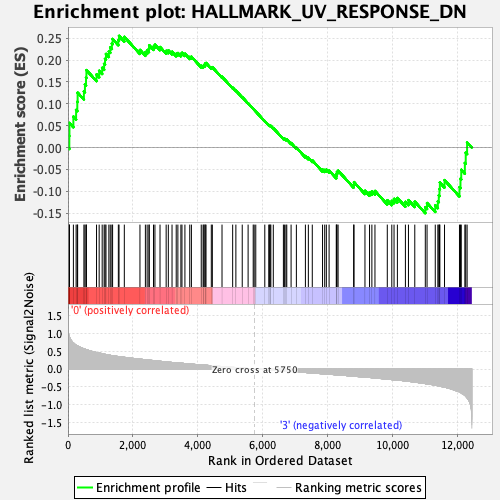
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.25547063 |
| Normalized Enrichment Score (NES) | 1.219199 |
| Nominal p-value | 0.07968128 |
| FDR q-value | 0.7799425 |
| FWER p-Value | 0.904 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmpr1a | 41 | 0.907 | 0.0267 | Yes |
| 2 | Pten | 45 | 0.905 | 0.0563 | Yes |
| 3 | Prdm2 | 166 | 0.735 | 0.0708 | Yes |
| 4 | Pex14 | 250 | 0.668 | 0.0862 | Yes |
| 5 | Gja1 | 287 | 0.653 | 0.1048 | Yes |
| 6 | Wdr37 | 297 | 0.646 | 0.1254 | Yes |
| 7 | Rasa2 | 490 | 0.567 | 0.1286 | Yes |
| 8 | Cdkn1b | 522 | 0.558 | 0.1445 | Yes |
| 9 | Notch2 | 556 | 0.549 | 0.1600 | Yes |
| 10 | Insig1 | 568 | 0.545 | 0.1771 | Yes |
| 11 | Mapk14 | 879 | 0.463 | 0.1673 | Yes |
| 12 | Bhlhe40 | 960 | 0.451 | 0.1757 | Yes |
| 13 | Mgmt | 1053 | 0.431 | 0.1825 | Yes |
| 14 | Smad7 | 1113 | 0.420 | 0.1915 | Yes |
| 15 | Apbb2 | 1141 | 0.412 | 0.2030 | Yes |
| 16 | Acvr2a | 1170 | 0.407 | 0.2141 | Yes |
| 17 | Slc7a1 | 1259 | 0.391 | 0.2199 | Yes |
| 18 | Arhgef9 | 1302 | 0.385 | 0.2292 | Yes |
| 19 | Nrp1 | 1352 | 0.377 | 0.2377 | Yes |
| 20 | Ythdc1 | 1374 | 0.374 | 0.2484 | Yes |
| 21 | Togaram1 | 1551 | 0.349 | 0.2456 | Yes |
| 22 | Vldlr | 1572 | 0.347 | 0.2555 | Yes |
| 23 | Dusp1 | 1735 | 0.328 | 0.2532 | No |
| 24 | Add3 | 2217 | 0.276 | 0.2233 | No |
| 25 | Atp2c1 | 2384 | 0.259 | 0.2184 | No |
| 26 | Atrn | 2438 | 0.254 | 0.2225 | No |
| 27 | Rbpms | 2494 | 0.250 | 0.2263 | No |
| 28 | Cdc42bpa | 2503 | 0.249 | 0.2339 | No |
| 29 | Prkca | 2634 | 0.237 | 0.2312 | No |
| 30 | Mgll | 2678 | 0.233 | 0.2354 | No |
| 31 | Anxa2 | 2835 | 0.218 | 0.2299 | No |
| 32 | Kit | 3024 | 0.199 | 0.2212 | No |
| 33 | Lamc1 | 3091 | 0.193 | 0.2222 | No |
| 34 | Sfmbt1 | 3205 | 0.184 | 0.2191 | No |
| 35 | Fyn | 3334 | 0.176 | 0.2146 | No |
| 36 | Sdc2 | 3385 | 0.170 | 0.2162 | No |
| 37 | Syne1 | 3478 | 0.163 | 0.2141 | No |
| 38 | Phf3 | 3511 | 0.161 | 0.2168 | No |
| 39 | Mios | 3602 | 0.154 | 0.2146 | No |
| 40 | Plcb4 | 3751 | 0.141 | 0.2073 | No |
| 41 | Celf2 | 3802 | 0.138 | 0.2078 | No |
| 42 | Ddah1 | 4105 | 0.114 | 0.1870 | No |
| 43 | Dyrk1a | 4162 | 0.112 | 0.1862 | No |
| 44 | Scaf8 | 4184 | 0.110 | 0.1882 | No |
| 45 | Tgfbr2 | 4222 | 0.107 | 0.1887 | No |
| 46 | Atrx | 4223 | 0.107 | 0.1922 | No |
| 47 | Inpp4b | 4252 | 0.105 | 0.1934 | No |
| 48 | Pdgfrb | 4415 | 0.092 | 0.1833 | No |
| 49 | Dbp | 4447 | 0.089 | 0.1838 | No |
| 50 | Pdlim5 | 4745 | 0.067 | 0.1619 | No |
| 51 | Dmac2l | 5073 | 0.047 | 0.1370 | No |
| 52 | Smad3 | 5172 | 0.039 | 0.1303 | No |
| 53 | Dlg1 | 5368 | 0.026 | 0.1153 | No |
| 54 | Atp2b4 | 5549 | 0.014 | 0.1012 | No |
| 55 | Nipbl | 5706 | 0.002 | 0.0886 | No |
| 56 | Bckdhb | 5736 | 0.001 | 0.0863 | No |
| 57 | Runx1 | 5785 | -0.000 | 0.0824 | No |
| 58 | Ica1 | 6063 | -0.018 | 0.0606 | No |
| 59 | Atp2b1 | 6185 | -0.025 | 0.0516 | No |
| 60 | Atxn1 | 6212 | -0.027 | 0.0504 | No |
| 61 | Ltbp1 | 6227 | -0.028 | 0.0502 | No |
| 62 | Lpar1 | 6249 | -0.029 | 0.0494 | No |
| 63 | Abcc1 | 6325 | -0.035 | 0.0445 | No |
| 64 | Anxa4 | 6636 | -0.055 | 0.0212 | No |
| 65 | Spop | 6667 | -0.056 | 0.0206 | No |
| 66 | Itgb3 | 6713 | -0.058 | 0.0189 | No |
| 67 | Prkce | 6740 | -0.060 | 0.0188 | No |
| 68 | Igf1r | 6873 | -0.069 | 0.0103 | No |
| 69 | Ldlr | 7036 | -0.079 | -0.0002 | No |
| 70 | Akt3 | 7314 | -0.097 | -0.0194 | No |
| 71 | Adgrl2 | 7405 | -0.104 | -0.0233 | No |
| 72 | Ptpn21 | 7526 | -0.112 | -0.0293 | No |
| 73 | Vav2 | 7842 | -0.134 | -0.0504 | No |
| 74 | Pik3cd | 7909 | -0.139 | -0.0512 | No |
| 75 | Kalrn | 7959 | -0.142 | -0.0505 | No |
| 76 | Pik3r3 | 8044 | -0.148 | -0.0524 | No |
| 77 | Zmiz1 | 8265 | -0.163 | -0.0649 | No |
| 78 | Grk5 | 8274 | -0.164 | -0.0601 | No |
| 79 | Pias3 | 8287 | -0.164 | -0.0557 | No |
| 80 | Met | 8321 | -0.166 | -0.0528 | No |
| 81 | Mta1 | 8800 | -0.202 | -0.0849 | No |
| 82 | Synj2 | 8812 | -0.203 | -0.0791 | No |
| 83 | Cited2 | 9149 | -0.227 | -0.0988 | No |
| 84 | Cdk13 | 9290 | -0.238 | -0.1023 | No |
| 85 | Tfpi | 9362 | -0.244 | -0.1000 | No |
| 86 | Aggf1 | 9457 | -0.253 | -0.0993 | No |
| 87 | Map2k5 | 9837 | -0.286 | -0.1206 | No |
| 88 | Rxra | 9976 | -0.298 | -0.1219 | No |
| 89 | Nr3c1 | 10045 | -0.305 | -0.1174 | No |
| 90 | Gcnt1 | 10148 | -0.312 | -0.1153 | No |
| 91 | Sipa1l1 | 10395 | -0.336 | -0.1242 | No |
| 92 | Tjp1 | 10489 | -0.346 | -0.1203 | No |
| 93 | Cap2 | 10684 | -0.370 | -0.1238 | No |
| 94 | Nfkb1 | 11008 | -0.411 | -0.1364 | No |
| 95 | Nfib | 11062 | -0.418 | -0.1269 | No |
| 96 | Sri | 11315 | -0.458 | -0.1322 | No |
| 97 | Mrps31 | 11395 | -0.474 | -0.1230 | No |
| 98 | Tgfbr3 | 11424 | -0.480 | -0.1094 | No |
| 99 | Cdon | 11445 | -0.483 | -0.0950 | No |
| 100 | Nek7 | 11457 | -0.484 | -0.0800 | No |
| 101 | Prkar2b | 11600 | -0.513 | -0.0745 | No |
| 102 | Magi2 | 12061 | -0.643 | -0.0906 | No |
| 103 | Nr1d2 | 12092 | -0.662 | -0.0712 | No |
| 104 | Tent4a | 12116 | -0.672 | -0.0508 | No |
| 105 | Myc | 12227 | -0.743 | -0.0352 | No |
| 106 | Cacna1a | 12254 | -0.773 | -0.0118 | No |
| 107 | Slc22a18 | 12296 | -0.813 | 0.0118 | No |