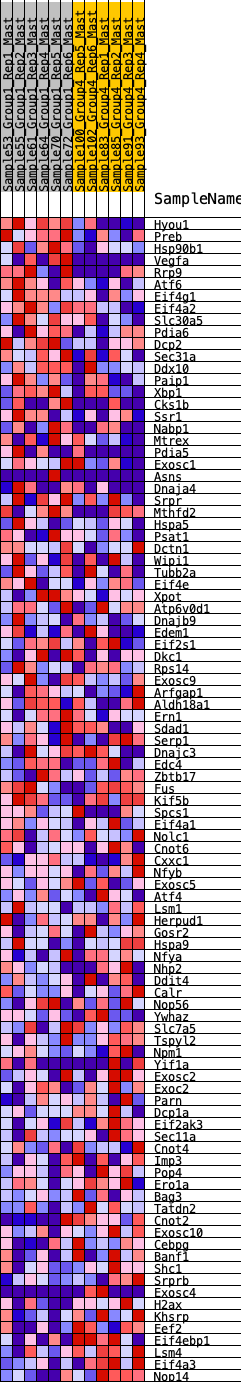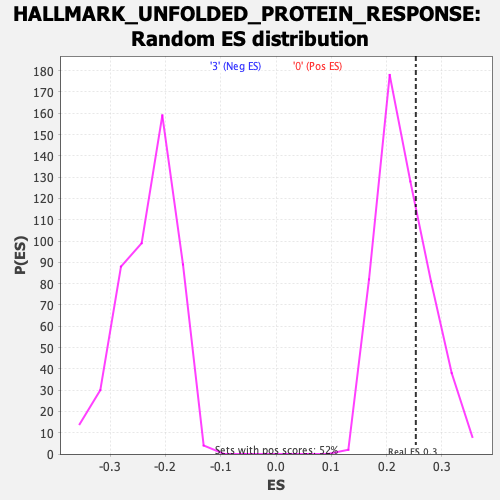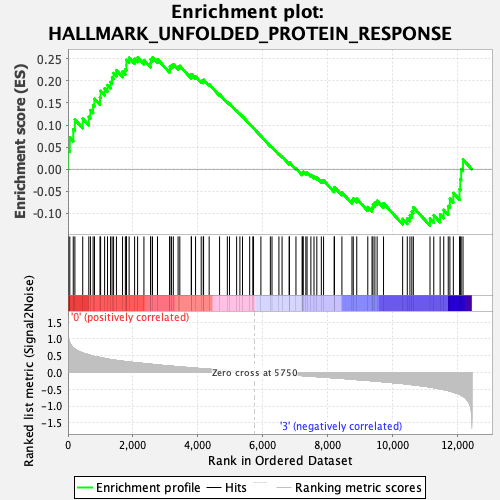
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.25307238 |
| Normalized Enrichment Score (NES) | 1.0895774 |
| Nominal p-value | 0.311412 |
| FDR q-value | 0.92891216 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hyou1 | 6 | 1.169 | 0.0430 | Yes |
| 2 | Preb | 55 | 0.877 | 0.0717 | Yes |
| 3 | Hsp90b1 | 162 | 0.737 | 0.0905 | Yes |
| 4 | Vegfa | 212 | 0.695 | 0.1123 | Yes |
| 5 | Rrp9 | 454 | 0.581 | 0.1144 | Yes |
| 6 | Atf6 | 641 | 0.520 | 0.1187 | Yes |
| 7 | Eif4g1 | 691 | 0.509 | 0.1337 | Yes |
| 8 | Eif4a2 | 773 | 0.488 | 0.1452 | Yes |
| 9 | Slc30a5 | 816 | 0.475 | 0.1595 | Yes |
| 10 | Pdia6 | 986 | 0.446 | 0.1624 | Yes |
| 11 | Dcp2 | 1005 | 0.441 | 0.1773 | Yes |
| 12 | Sec31a | 1128 | 0.415 | 0.1828 | Yes |
| 13 | Ddx10 | 1217 | 0.398 | 0.1905 | Yes |
| 14 | Paip1 | 1311 | 0.384 | 0.1972 | Yes |
| 15 | Xbp1 | 1358 | 0.377 | 0.2075 | Yes |
| 16 | Cks1b | 1404 | 0.370 | 0.2176 | Yes |
| 17 | Ssr1 | 1492 | 0.358 | 0.2238 | Yes |
| 18 | Nabp1 | 1680 | 0.334 | 0.2211 | Yes |
| 19 | Mtrex | 1767 | 0.325 | 0.2262 | Yes |
| 20 | Pdia5 | 1799 | 0.321 | 0.2357 | Yes |
| 21 | Exosc1 | 1800 | 0.321 | 0.2476 | Yes |
| 22 | Asns | 1883 | 0.311 | 0.2525 | Yes |
| 23 | Dnaja4 | 2052 | 0.293 | 0.2498 | Yes |
| 24 | Srpr | 2144 | 0.283 | 0.2529 | Yes |
| 25 | Mthfd2 | 2339 | 0.265 | 0.2471 | Yes |
| 26 | Hspa5 | 2544 | 0.245 | 0.2397 | Yes |
| 27 | Psat1 | 2548 | 0.245 | 0.2485 | Yes |
| 28 | Dctn1 | 2603 | 0.240 | 0.2531 | Yes |
| 29 | Wipi1 | 2757 | 0.224 | 0.2490 | No |
| 30 | Tubb2a | 3131 | 0.189 | 0.2258 | No |
| 31 | Eif4e | 3139 | 0.189 | 0.2323 | No |
| 32 | Xpot | 3190 | 0.185 | 0.2351 | No |
| 33 | Atp6v0d1 | 3245 | 0.181 | 0.2375 | No |
| 34 | Dnajb9 | 3392 | 0.170 | 0.2319 | No |
| 35 | Edem1 | 3445 | 0.165 | 0.2339 | No |
| 36 | Eif2s1 | 3797 | 0.138 | 0.2106 | No |
| 37 | Dkc1 | 3804 | 0.138 | 0.2152 | No |
| 38 | Rps14 | 3930 | 0.127 | 0.2098 | No |
| 39 | Exosc9 | 4104 | 0.114 | 0.2000 | No |
| 40 | Arfgap1 | 4167 | 0.112 | 0.1992 | No |
| 41 | Aldh18a1 | 4177 | 0.111 | 0.2025 | No |
| 42 | Ern1 | 4349 | 0.098 | 0.1923 | No |
| 43 | Sdad1 | 4670 | 0.073 | 0.1691 | No |
| 44 | Serp1 | 4909 | 0.056 | 0.1519 | No |
| 45 | Dnajc3 | 4976 | 0.052 | 0.1485 | No |
| 46 | Edc4 | 5195 | 0.037 | 0.1323 | No |
| 47 | Zbtb17 | 5296 | 0.030 | 0.1253 | No |
| 48 | Fus | 5376 | 0.025 | 0.1198 | No |
| 49 | Kif5b | 5597 | 0.010 | 0.1024 | No |
| 50 | Spcs1 | 5694 | 0.003 | 0.0947 | No |
| 51 | Eif4a1 | 5716 | 0.002 | 0.0931 | No |
| 52 | Nolc1 | 5943 | -0.011 | 0.0752 | No |
| 53 | Cnot6 | 6234 | -0.029 | 0.0528 | No |
| 54 | Cxxc1 | 6285 | -0.032 | 0.0499 | No |
| 55 | Nfyb | 6498 | -0.046 | 0.0345 | No |
| 56 | Exosc5 | 6595 | -0.052 | 0.0286 | No |
| 57 | Atf4 | 6813 | -0.065 | 0.0135 | No |
| 58 | Lsm1 | 6823 | -0.066 | 0.0152 | No |
| 59 | Herpud1 | 7023 | -0.078 | 0.0020 | No |
| 60 | Gosr2 | 7213 | -0.091 | -0.0099 | No |
| 61 | Hspa9 | 7228 | -0.093 | -0.0076 | No |
| 62 | Nfya | 7248 | -0.094 | -0.0057 | No |
| 63 | Nhp2 | 7328 | -0.099 | -0.0084 | No |
| 64 | Ddit4 | 7365 | -0.101 | -0.0076 | No |
| 65 | Calr | 7483 | -0.109 | -0.0130 | No |
| 66 | Nop56 | 7579 | -0.116 | -0.0164 | No |
| 67 | Ywhaz | 7666 | -0.122 | -0.0188 | No |
| 68 | Slc7a5 | 7805 | -0.131 | -0.0251 | No |
| 69 | Tspyl2 | 7874 | -0.136 | -0.0255 | No |
| 70 | Npm1 | 8200 | -0.158 | -0.0460 | No |
| 71 | Yif1a | 8209 | -0.159 | -0.0407 | No |
| 72 | Exosc2 | 8439 | -0.175 | -0.0527 | No |
| 73 | Exoc2 | 8749 | -0.198 | -0.0704 | No |
| 74 | Parn | 8789 | -0.201 | -0.0661 | No |
| 75 | Dcp1a | 8896 | -0.209 | -0.0669 | No |
| 76 | Eif2ak3 | 9236 | -0.233 | -0.0857 | No |
| 77 | Sec11a | 9367 | -0.244 | -0.0872 | No |
| 78 | Cnot4 | 9396 | -0.247 | -0.0803 | No |
| 79 | Imp3 | 9455 | -0.253 | -0.0756 | No |
| 80 | Pop4 | 9524 | -0.259 | -0.0714 | No |
| 81 | Ero1a | 9721 | -0.275 | -0.0771 | No |
| 82 | Bag3 | 10310 | -0.326 | -0.1126 | No |
| 83 | Tatdn2 | 10453 | -0.342 | -0.1114 | No |
| 84 | Cnot2 | 10534 | -0.352 | -0.1048 | No |
| 85 | Exosc10 | 10593 | -0.360 | -0.0961 | No |
| 86 | Cebpg | 10640 | -0.365 | -0.0862 | No |
| 87 | Banf1 | 11153 | -0.430 | -0.1117 | No |
| 88 | Shc1 | 11269 | -0.450 | -0.1043 | No |
| 89 | Srprb | 11465 | -0.486 | -0.1021 | No |
| 90 | Exosc4 | 11576 | -0.508 | -0.0921 | No |
| 91 | H2ax | 11715 | -0.536 | -0.0833 | No |
| 92 | Khsrp | 11766 | -0.550 | -0.0670 | No |
| 93 | Eef2 | 11872 | -0.583 | -0.0538 | No |
| 94 | Eif4ebp1 | 12063 | -0.643 | -0.0453 | No |
| 95 | Lsm4 | 12088 | -0.660 | -0.0227 | No |
| 96 | Eif4a3 | 12113 | -0.672 | 0.0003 | No |
| 97 | Nop14 | 12170 | -0.705 | 0.0220 | No |