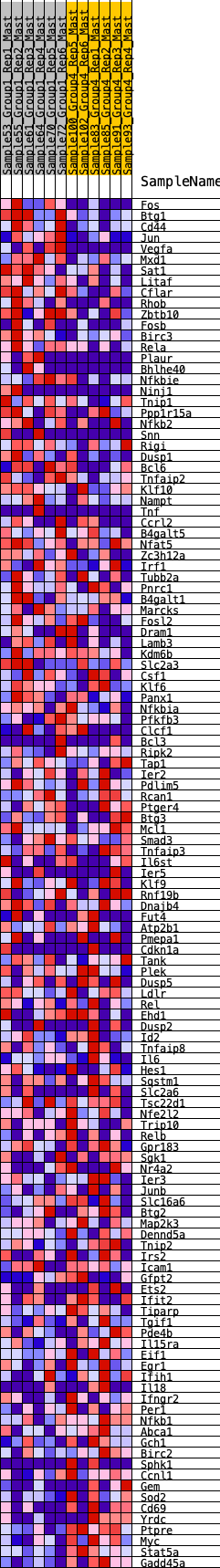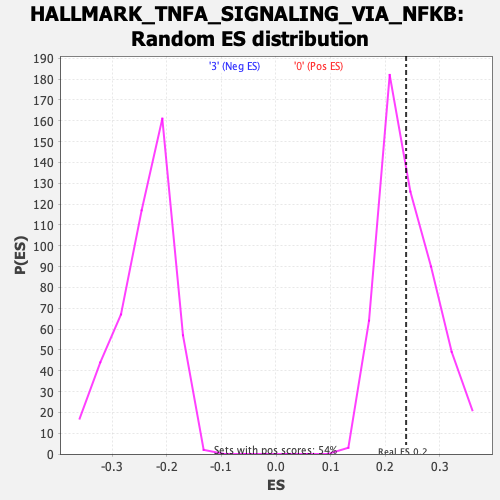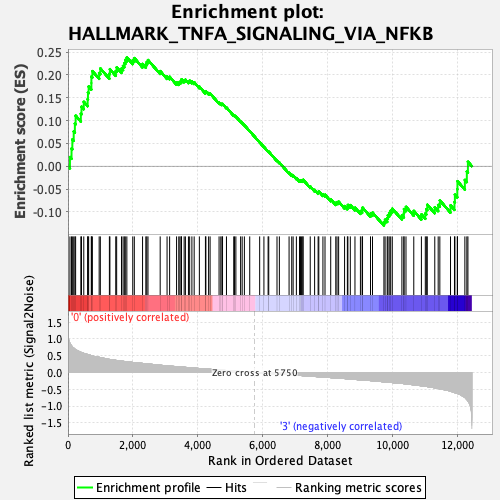
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.23829554 |
| Normalized Enrichment Score (NES) | 0.9858591 |
| Nominal p-value | 0.46542057 |
| FDR q-value | 0.7337759 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fos | 60 | 0.871 | 0.0201 | Yes |
| 2 | Btg1 | 109 | 0.790 | 0.0388 | Yes |
| 3 | Cd44 | 134 | 0.761 | 0.0586 | Yes |
| 4 | Jun | 176 | 0.726 | 0.0761 | Yes |
| 5 | Vegfa | 212 | 0.695 | 0.0931 | Yes |
| 6 | Mxd1 | 235 | 0.674 | 0.1106 | Yes |
| 7 | Sat1 | 396 | 0.601 | 0.1149 | Yes |
| 8 | Litaf | 416 | 0.595 | 0.1303 | Yes |
| 9 | Cflar | 486 | 0.569 | 0.1410 | Yes |
| 10 | Rhob | 605 | 0.535 | 0.1467 | Yes |
| 11 | Zbtb10 | 615 | 0.530 | 0.1612 | Yes |
| 12 | Fosb | 636 | 0.523 | 0.1745 | Yes |
| 13 | Birc3 | 720 | 0.501 | 0.1821 | Yes |
| 14 | Rela | 721 | 0.500 | 0.1965 | Yes |
| 15 | Plaur | 751 | 0.493 | 0.2082 | Yes |
| 16 | Bhlhe40 | 960 | 0.451 | 0.2043 | Yes |
| 17 | Nfkbie | 996 | 0.444 | 0.2141 | Yes |
| 18 | Ninj1 | 1275 | 0.389 | 0.2027 | Yes |
| 19 | Tnip1 | 1291 | 0.387 | 0.2125 | Yes |
| 20 | Ppp1r15a | 1471 | 0.361 | 0.2083 | Yes |
| 21 | Nfkb2 | 1499 | 0.357 | 0.2164 | Yes |
| 22 | Snn | 1648 | 0.337 | 0.2140 | Yes |
| 23 | Rigi | 1696 | 0.332 | 0.2197 | Yes |
| 24 | Dusp1 | 1735 | 0.328 | 0.2260 | Yes |
| 25 | Bcl6 | 1765 | 0.325 | 0.2330 | Yes |
| 26 | Tnfaip2 | 1813 | 0.320 | 0.2383 | Yes |
| 27 | Klf10 | 1995 | 0.299 | 0.2322 | No |
| 28 | Nampt | 2043 | 0.295 | 0.2368 | No |
| 29 | Tnf | 2297 | 0.269 | 0.2240 | No |
| 30 | Ccrl2 | 2398 | 0.258 | 0.2232 | No |
| 31 | B4galt5 | 2427 | 0.255 | 0.2282 | No |
| 32 | Nfat5 | 2464 | 0.252 | 0.2325 | No |
| 33 | Zc3h12a | 2840 | 0.217 | 0.2083 | No |
| 34 | Irf1 | 3052 | 0.196 | 0.1967 | No |
| 35 | Tubb2a | 3131 | 0.189 | 0.1958 | No |
| 36 | Pnrc1 | 3345 | 0.175 | 0.1835 | No |
| 37 | B4galt1 | 3413 | 0.168 | 0.1829 | No |
| 38 | Marcks | 3448 | 0.165 | 0.1848 | No |
| 39 | Fosl2 | 3485 | 0.163 | 0.1866 | No |
| 40 | Dram1 | 3497 | 0.162 | 0.1903 | No |
| 41 | Lamb3 | 3578 | 0.155 | 0.1883 | No |
| 42 | Kdm6b | 3617 | 0.152 | 0.1895 | No |
| 43 | Slc2a3 | 3711 | 0.145 | 0.1861 | No |
| 44 | Csf1 | 3748 | 0.141 | 0.1873 | No |
| 45 | Klf6 | 3817 | 0.136 | 0.1857 | No |
| 46 | Panx1 | 3887 | 0.131 | 0.1838 | No |
| 47 | Nfkbia | 4047 | 0.118 | 0.1743 | No |
| 48 | Pfkfb3 | 4234 | 0.106 | 0.1622 | No |
| 49 | Clcf1 | 4248 | 0.105 | 0.1642 | No |
| 50 | Bcl3 | 4332 | 0.098 | 0.1602 | No |
| 51 | Ripk2 | 4378 | 0.095 | 0.1593 | No |
| 52 | Tap1 | 4654 | 0.075 | 0.1391 | No |
| 53 | Ier2 | 4708 | 0.070 | 0.1368 | No |
| 54 | Pdlim5 | 4745 | 0.067 | 0.1358 | No |
| 55 | Rcan1 | 4761 | 0.066 | 0.1365 | No |
| 56 | Ptger4 | 4882 | 0.058 | 0.1284 | No |
| 57 | Btg3 | 5106 | 0.045 | 0.1116 | No |
| 58 | Mcl1 | 5131 | 0.042 | 0.1108 | No |
| 59 | Smad3 | 5172 | 0.039 | 0.1087 | No |
| 60 | Tnfaip3 | 5318 | 0.028 | 0.0978 | No |
| 61 | Il6st | 5366 | 0.026 | 0.0947 | No |
| 62 | Ier5 | 5434 | 0.021 | 0.0898 | No |
| 63 | Klf9 | 5599 | 0.010 | 0.0768 | No |
| 64 | Rnf19b | 5905 | -0.009 | 0.0523 | No |
| 65 | Dnajb4 | 6036 | -0.016 | 0.0422 | No |
| 66 | Fut4 | 6165 | -0.024 | 0.0325 | No |
| 67 | Atp2b1 | 6185 | -0.025 | 0.0317 | No |
| 68 | Pmepa1 | 6439 | -0.043 | 0.0124 | No |
| 69 | Cdkn1a | 6512 | -0.047 | 0.0079 | No |
| 70 | Tank | 6811 | -0.065 | -0.0144 | No |
| 71 | Plek | 6893 | -0.070 | -0.0190 | No |
| 72 | Dusp5 | 6936 | -0.073 | -0.0203 | No |
| 73 | Ldlr | 7036 | -0.079 | -0.0261 | No |
| 74 | Rel | 7133 | -0.085 | -0.0315 | No |
| 75 | Ehd1 | 7164 | -0.087 | -0.0314 | No |
| 76 | Dusp2 | 7192 | -0.090 | -0.0310 | No |
| 77 | Id2 | 7232 | -0.093 | -0.0315 | No |
| 78 | Tnfaip8 | 7244 | -0.094 | -0.0298 | No |
| 79 | Il6 | 7463 | -0.108 | -0.0444 | No |
| 80 | Hes1 | 7598 | -0.117 | -0.0519 | No |
| 81 | Sqstm1 | 7706 | -0.125 | -0.0570 | No |
| 82 | Slc2a6 | 7727 | -0.126 | -0.0550 | No |
| 83 | Tsc22d1 | 7854 | -0.134 | -0.0614 | No |
| 84 | Nfe2l2 | 7918 | -0.139 | -0.0625 | No |
| 85 | Trip10 | 8094 | -0.151 | -0.0724 | No |
| 86 | Relb | 8250 | -0.162 | -0.0803 | No |
| 87 | Gpr183 | 8288 | -0.164 | -0.0786 | No |
| 88 | Sgk1 | 8338 | -0.168 | -0.0778 | No |
| 89 | Nr4a2 | 8521 | -0.180 | -0.0875 | No |
| 90 | Ier3 | 8607 | -0.187 | -0.0890 | No |
| 91 | Junb | 8623 | -0.188 | -0.0848 | No |
| 92 | Slc16a6 | 8698 | -0.194 | -0.0853 | No |
| 93 | Btg2 | 8841 | -0.206 | -0.0910 | No |
| 94 | Map2k3 | 9009 | -0.217 | -0.0983 | No |
| 95 | Dennd5a | 9061 | -0.220 | -0.0961 | No |
| 96 | Tnip2 | 9074 | -0.221 | -0.0908 | No |
| 97 | Irs2 | 9320 | -0.241 | -0.1038 | No |
| 98 | Icam1 | 9381 | -0.246 | -0.1016 | No |
| 99 | Gfpt2 | 9733 | -0.276 | -0.1222 | No |
| 100 | Ets2 | 9769 | -0.279 | -0.1171 | No |
| 101 | Ifit2 | 9834 | -0.285 | -0.1141 | No |
| 102 | Tiparp | 9855 | -0.287 | -0.1075 | No |
| 103 | Tgif1 | 9896 | -0.291 | -0.1024 | No |
| 104 | Pde4b | 9941 | -0.295 | -0.0976 | No |
| 105 | Il15ra | 9991 | -0.300 | -0.0930 | No |
| 106 | Eif1 | 10287 | -0.325 | -0.1076 | No |
| 107 | Egr1 | 10348 | -0.331 | -0.1030 | No |
| 108 | Ifih1 | 10352 | -0.331 | -0.0938 | No |
| 109 | Il18 | 10413 | -0.337 | -0.0890 | No |
| 110 | Ifngr2 | 10653 | -0.367 | -0.0979 | No |
| 111 | Per1 | 10888 | -0.394 | -0.1056 | No |
| 112 | Nfkb1 | 11008 | -0.411 | -0.1035 | No |
| 113 | Abca1 | 11040 | -0.416 | -0.0941 | No |
| 114 | Gch1 | 11069 | -0.419 | -0.0844 | No |
| 115 | Birc2 | 11300 | -0.456 | -0.0900 | No |
| 116 | Sphk1 | 11405 | -0.476 | -0.0849 | No |
| 117 | Ccnl1 | 11456 | -0.484 | -0.0751 | No |
| 118 | Gem | 11785 | -0.555 | -0.0858 | No |
| 119 | Sod2 | 11907 | -0.595 | -0.0786 | No |
| 120 | Cd69 | 11920 | -0.600 | -0.0624 | No |
| 121 | Yrdc | 11994 | -0.624 | -0.0505 | No |
| 122 | Ptpre | 11999 | -0.625 | -0.0329 | No |
| 123 | Myc | 12227 | -0.743 | -0.0300 | No |
| 124 | Stat5a | 12288 | -0.806 | -0.0118 | No |
| 125 | Gadd45a | 12321 | -0.845 | 0.0097 | No |