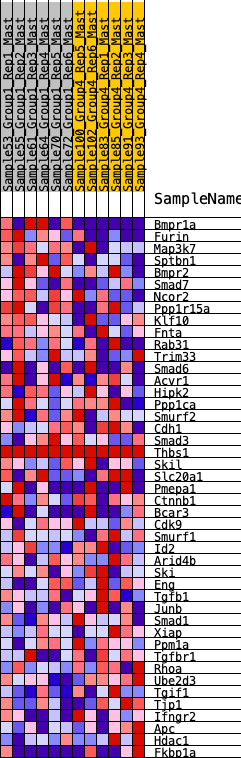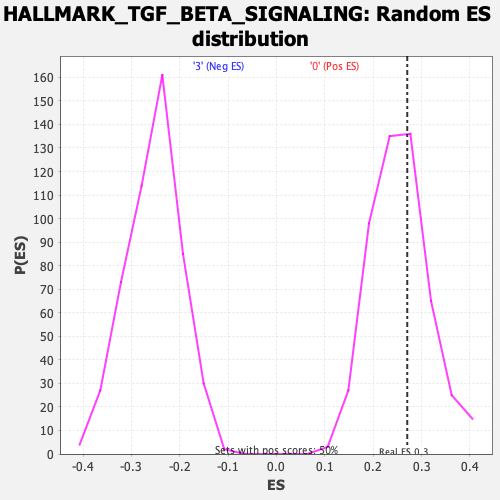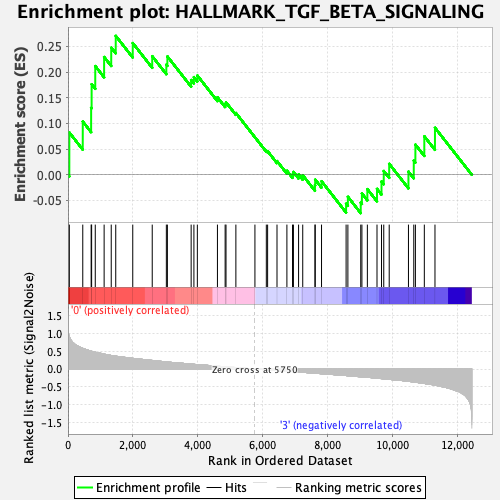
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.27052203 |
| Normalized Enrichment Score (NES) | 1.067339 |
| Nominal p-value | 0.3531746 |
| FDR q-value | 0.90808123 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmpr1a | 41 | 0.907 | 0.0822 | Yes |
| 2 | Furin | 455 | 0.580 | 0.1036 | Yes |
| 3 | Map3k7 | 713 | 0.503 | 0.1304 | Yes |
| 4 | Sptbn1 | 728 | 0.499 | 0.1763 | Yes |
| 5 | Bmpr2 | 840 | 0.471 | 0.2117 | Yes |
| 6 | Smad7 | 1113 | 0.420 | 0.2294 | Yes |
| 7 | Ncor2 | 1332 | 0.380 | 0.2476 | Yes |
| 8 | Ppp1r15a | 1471 | 0.361 | 0.2705 | Yes |
| 9 | Klf10 | 1995 | 0.299 | 0.2566 | No |
| 10 | Fnta | 2595 | 0.240 | 0.2309 | No |
| 11 | Rab31 | 3030 | 0.198 | 0.2146 | No |
| 12 | Trim33 | 3062 | 0.195 | 0.2305 | No |
| 13 | Smad6 | 3794 | 0.138 | 0.1846 | No |
| 14 | Acvr1 | 3879 | 0.131 | 0.1902 | No |
| 15 | Hipk2 | 3985 | 0.123 | 0.1933 | No |
| 16 | Ppp1ca | 4604 | 0.079 | 0.1509 | No |
| 17 | Smurf2 | 4841 | 0.061 | 0.1376 | No |
| 18 | Cdh1 | 4868 | 0.059 | 0.1411 | No |
| 19 | Smad3 | 5172 | 0.039 | 0.1203 | No |
| 20 | Thbs1 | 5759 | 0.000 | 0.0730 | No |
| 21 | Skil | 6110 | -0.021 | 0.0468 | No |
| 22 | Slc20a1 | 6150 | -0.024 | 0.0459 | No |
| 23 | Pmepa1 | 6439 | -0.043 | 0.0266 | No |
| 24 | Ctnnb1 | 6744 | -0.060 | 0.0078 | No |
| 25 | Bcar3 | 6917 | -0.072 | 0.0007 | No |
| 26 | Cdk9 | 6945 | -0.074 | 0.0055 | No |
| 27 | Smurf1 | 7104 | -0.083 | 0.0006 | No |
| 28 | Id2 | 7232 | -0.093 | -0.0009 | No |
| 29 | Arid4b | 7609 | -0.118 | -0.0201 | No |
| 30 | Ski | 7616 | -0.119 | -0.0094 | No |
| 31 | Eng | 7811 | -0.132 | -0.0126 | No |
| 32 | Tgfb1 | 8568 | -0.184 | -0.0563 | No |
| 33 | Junb | 8623 | -0.188 | -0.0429 | No |
| 34 | Smad1 | 9019 | -0.217 | -0.0543 | No |
| 35 | Xiap | 9055 | -0.220 | -0.0364 | No |
| 36 | Ppm1a | 9223 | -0.232 | -0.0280 | No |
| 37 | Tgfbr1 | 9520 | -0.259 | -0.0275 | No |
| 38 | Rhoa | 9659 | -0.270 | -0.0131 | No |
| 39 | Ube2d3 | 9728 | -0.275 | 0.0073 | No |
| 40 | Tgif1 | 9896 | -0.291 | 0.0213 | No |
| 41 | Tjp1 | 10489 | -0.346 | 0.0061 | No |
| 42 | Ifngr2 | 10653 | -0.367 | 0.0276 | No |
| 43 | Apc | 10703 | -0.372 | 0.0587 | No |
| 44 | Hdac1 | 10978 | -0.406 | 0.0749 | No |
| 45 | Fkbp1a | 11306 | -0.457 | 0.0916 | No |