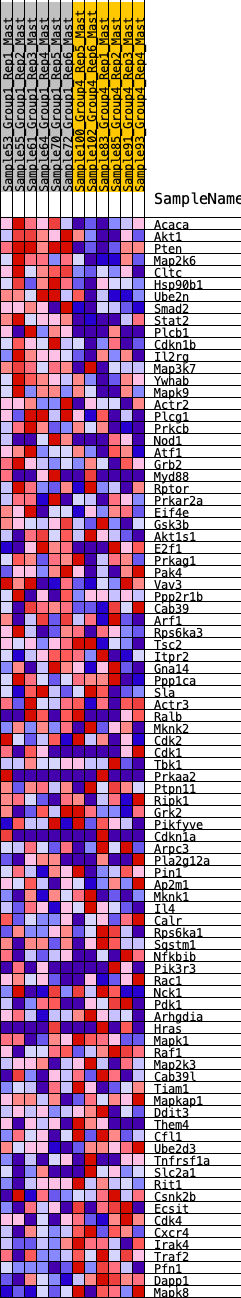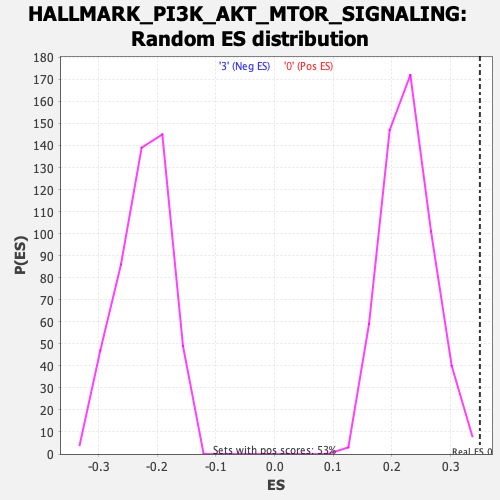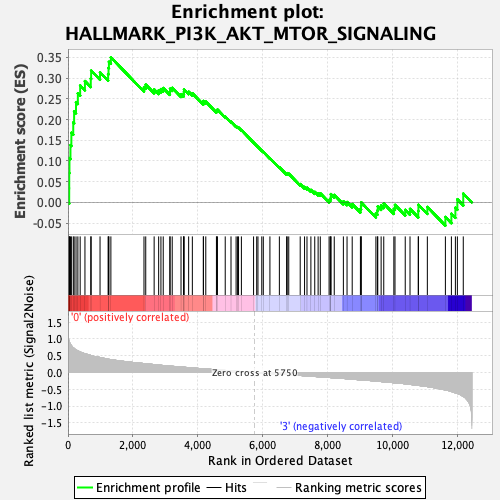
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.34989035 |
| Normalized Enrichment Score (NES) | 1.536947 |
| Nominal p-value | 0.003773585 |
| FDR q-value | 0.21310481 |
| FWER p-Value | 0.207 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acaca | 37 | 0.922 | 0.0341 | Yes |
| 2 | Akt1 | 38 | 0.920 | 0.0710 | Yes |
| 3 | Pten | 45 | 0.905 | 0.1069 | Yes |
| 4 | Map2k6 | 76 | 0.838 | 0.1382 | Yes |
| 5 | Cltc | 104 | 0.796 | 0.1680 | Yes |
| 6 | Hsp90b1 | 162 | 0.737 | 0.1930 | Yes |
| 7 | Ube2n | 189 | 0.714 | 0.2196 | Yes |
| 8 | Smad2 | 247 | 0.670 | 0.2419 | Yes |
| 9 | Stat2 | 304 | 0.643 | 0.2632 | Yes |
| 10 | Plcb1 | 375 | 0.612 | 0.2822 | Yes |
| 11 | Cdkn1b | 522 | 0.558 | 0.2928 | Yes |
| 12 | Il2rg | 702 | 0.507 | 0.2987 | Yes |
| 13 | Map3k7 | 713 | 0.503 | 0.3181 | Yes |
| 14 | Ywhab | 987 | 0.445 | 0.3139 | Yes |
| 15 | Mapk9 | 1236 | 0.395 | 0.3097 | Yes |
| 16 | Actr2 | 1249 | 0.393 | 0.3245 | Yes |
| 17 | Plcg1 | 1261 | 0.391 | 0.3393 | Yes |
| 18 | Prkcb | 1321 | 0.382 | 0.3499 | Yes |
| 19 | Nod1 | 2341 | 0.265 | 0.2781 | No |
| 20 | Atf1 | 2395 | 0.258 | 0.2841 | No |
| 21 | Grb2 | 2654 | 0.234 | 0.2727 | No |
| 22 | Myd88 | 2793 | 0.221 | 0.2704 | No |
| 23 | Rptor | 2863 | 0.214 | 0.2734 | No |
| 24 | Prkar2a | 2934 | 0.207 | 0.2760 | No |
| 25 | Eif4e | 3139 | 0.189 | 0.2671 | No |
| 26 | Gsk3b | 3144 | 0.188 | 0.2743 | No |
| 27 | Akt1s1 | 3211 | 0.184 | 0.2764 | No |
| 28 | E2f1 | 3482 | 0.163 | 0.2611 | No |
| 29 | Prkag1 | 3558 | 0.157 | 0.2613 | No |
| 30 | Pak4 | 3571 | 0.156 | 0.2666 | No |
| 31 | Vav3 | 3575 | 0.156 | 0.2726 | No |
| 32 | Ppp2r1b | 3717 | 0.144 | 0.2670 | No |
| 33 | Cab39 | 3831 | 0.135 | 0.2633 | No |
| 34 | Arf1 | 4170 | 0.111 | 0.2404 | No |
| 35 | Rps6ka3 | 4172 | 0.111 | 0.2448 | No |
| 36 | Tsc2 | 4245 | 0.106 | 0.2432 | No |
| 37 | Itpr2 | 4573 | 0.081 | 0.2200 | No |
| 38 | Gna14 | 4585 | 0.080 | 0.2223 | No |
| 39 | Ppp1ca | 4604 | 0.079 | 0.2240 | No |
| 40 | Sla | 4844 | 0.061 | 0.2071 | No |
| 41 | Actr3 | 5022 | 0.050 | 0.1948 | No |
| 42 | Ralb | 5179 | 0.038 | 0.1837 | No |
| 43 | Mknk2 | 5225 | 0.036 | 0.1815 | No |
| 44 | Cdk2 | 5254 | 0.034 | 0.1806 | No |
| 45 | Cdk1 | 5342 | 0.027 | 0.1746 | No |
| 46 | Tbk1 | 5717 | 0.002 | 0.1444 | No |
| 47 | Prkaa2 | 5814 | -0.002 | 0.1367 | No |
| 48 | Ptpn11 | 5852 | -0.005 | 0.1339 | No |
| 49 | Ripk1 | 5970 | -0.013 | 0.1250 | No |
| 50 | Grk2 | 6017 | -0.016 | 0.1219 | No |
| 51 | Pikfyve | 6221 | -0.028 | 0.1065 | No |
| 52 | Cdkn1a | 6512 | -0.047 | 0.0850 | No |
| 53 | Arpc3 | 6735 | -0.060 | 0.0694 | No |
| 54 | Pla2g12a | 6753 | -0.061 | 0.0705 | No |
| 55 | Pin1 | 6801 | -0.064 | 0.0693 | No |
| 56 | Ap2m1 | 7154 | -0.087 | 0.0442 | No |
| 57 | Mknk1 | 7288 | -0.096 | 0.0373 | No |
| 58 | Il4 | 7363 | -0.101 | 0.0354 | No |
| 59 | Calr | 7483 | -0.109 | 0.0302 | No |
| 60 | Rps6ka1 | 7602 | -0.118 | 0.0253 | No |
| 61 | Sqstm1 | 7706 | -0.125 | 0.0220 | No |
| 62 | Nfkbib | 7773 | -0.129 | 0.0219 | No |
| 63 | Pik3r3 | 8044 | -0.148 | 0.0060 | No |
| 64 | Rac1 | 8081 | -0.150 | 0.0091 | No |
| 65 | Nck1 | 8098 | -0.152 | 0.0139 | No |
| 66 | Pdk1 | 8099 | -0.152 | 0.0200 | No |
| 67 | Arhgdia | 8203 | -0.159 | 0.0180 | No |
| 68 | Hras | 8484 | -0.177 | 0.0025 | No |
| 69 | Mapk1 | 8597 | -0.186 | 0.0009 | No |
| 70 | Raf1 | 8756 | -0.198 | -0.0039 | No |
| 71 | Map2k3 | 9009 | -0.217 | -0.0156 | No |
| 72 | Cab39l | 9029 | -0.218 | -0.0084 | No |
| 73 | Tiam1 | 9033 | -0.218 | 0.0001 | No |
| 74 | Mapkap1 | 9486 | -0.256 | -0.0262 | No |
| 75 | Ddit3 | 9533 | -0.260 | -0.0195 | No |
| 76 | Them4 | 9544 | -0.260 | -0.0098 | No |
| 77 | Cfl1 | 9647 | -0.269 | -0.0072 | No |
| 78 | Ube2d3 | 9728 | -0.275 | -0.0026 | No |
| 79 | Tnfrsf1a | 10039 | -0.304 | -0.0155 | No |
| 80 | Slc2a1 | 10074 | -0.306 | -0.0060 | No |
| 81 | Rit1 | 10390 | -0.336 | -0.0180 | No |
| 82 | Csnk2b | 10537 | -0.353 | -0.0156 | No |
| 83 | Ecsit | 10791 | -0.383 | -0.0207 | No |
| 84 | Cdk4 | 10797 | -0.384 | -0.0057 | No |
| 85 | Cxcr4 | 11071 | -0.419 | -0.0110 | No |
| 86 | Irak4 | 11627 | -0.517 | -0.0351 | No |
| 87 | Traf2 | 11812 | -0.565 | -0.0273 | No |
| 88 | Pfn1 | 11935 | -0.606 | -0.0128 | No |
| 89 | Dapp1 | 11996 | -0.625 | 0.0074 | No |
| 90 | Mapk8 | 12178 | -0.710 | 0.0213 | No |