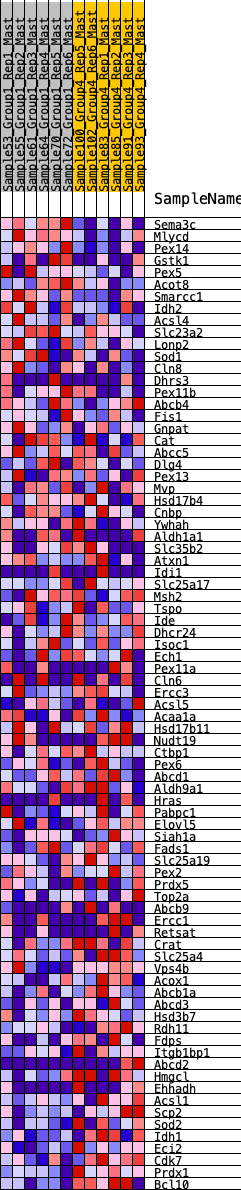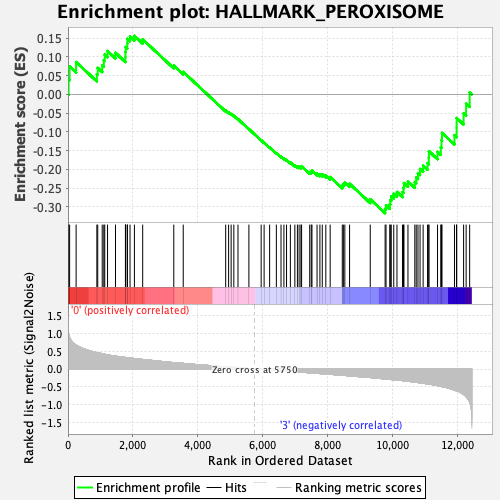
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.317078 |
| Normalized Enrichment Score (NES) | -1.3179893 |
| Nominal p-value | 0.06704981 |
| FDR q-value | 0.2906205 |
| FWER p-Value | 0.755 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sema3c | 19 | 1.001 | 0.0400 | No |
| 2 | Mlycd | 50 | 0.890 | 0.0746 | No |
| 3 | Pex14 | 250 | 0.668 | 0.0862 | No |
| 4 | Gstk1 | 889 | 0.461 | 0.0538 | No |
| 5 | Pex5 | 916 | 0.458 | 0.0707 | No |
| 6 | Acot8 | 1054 | 0.431 | 0.0775 | No |
| 7 | Smarcc1 | 1103 | 0.422 | 0.0912 | No |
| 8 | Idh2 | 1132 | 0.414 | 0.1061 | No |
| 9 | Acsl4 | 1218 | 0.397 | 0.1157 | No |
| 10 | Slc23a2 | 1463 | 0.363 | 0.1110 | No |
| 11 | Lonp2 | 1769 | 0.325 | 0.0998 | No |
| 12 | Sod1 | 1771 | 0.325 | 0.1132 | No |
| 13 | Cln8 | 1776 | 0.324 | 0.1264 | No |
| 14 | Dhrs3 | 1827 | 0.317 | 0.1355 | No |
| 15 | Pex11b | 1830 | 0.317 | 0.1485 | No |
| 16 | Abcb4 | 1911 | 0.308 | 0.1548 | No |
| 17 | Fis1 | 2046 | 0.294 | 0.1562 | No |
| 18 | Gnpat | 2301 | 0.269 | 0.1468 | No |
| 19 | Cat | 3259 | 0.180 | 0.0769 | No |
| 20 | Abcc5 | 3550 | 0.158 | 0.0600 | No |
| 21 | Dlg4 | 4858 | 0.060 | -0.0433 | No |
| 22 | Pex13 | 4948 | 0.054 | -0.0482 | No |
| 23 | Mvp | 5026 | 0.050 | -0.0524 | No |
| 24 | Hsd17b4 | 5109 | 0.044 | -0.0572 | No |
| 25 | Cnbp | 5237 | 0.035 | -0.0660 | No |
| 26 | Ywhah | 5573 | 0.012 | -0.0926 | No |
| 27 | Aldh1a1 | 5951 | -0.012 | -0.1226 | No |
| 28 | Slc35b2 | 6042 | -0.016 | -0.1292 | No |
| 29 | Atxn1 | 6212 | -0.027 | -0.1418 | No |
| 30 | Idi1 | 6419 | -0.041 | -0.1567 | No |
| 31 | Slc25a17 | 6563 | -0.051 | -0.1662 | No |
| 32 | Msh2 | 6652 | -0.055 | -0.1710 | No |
| 33 | Tspo | 6733 | -0.060 | -0.1750 | No |
| 34 | Ide | 6851 | -0.068 | -0.1817 | No |
| 35 | Dhcr24 | 6989 | -0.077 | -0.1896 | No |
| 36 | Isoc1 | 7068 | -0.081 | -0.1925 | No |
| 37 | Ech1 | 7120 | -0.084 | -0.1931 | No |
| 38 | Pex11a | 7176 | -0.088 | -0.1939 | No |
| 39 | Cln6 | 7197 | -0.090 | -0.1918 | No |
| 40 | Ercc3 | 7447 | -0.107 | -0.2075 | No |
| 41 | Acsl5 | 7494 | -0.110 | -0.2066 | No |
| 42 | Acaa1a | 7516 | -0.112 | -0.2037 | No |
| 43 | Hsd17b11 | 7673 | -0.122 | -0.2112 | No |
| 44 | Nudt19 | 7761 | -0.129 | -0.2129 | No |
| 45 | Ctbp1 | 7834 | -0.133 | -0.2132 | No |
| 46 | Pex6 | 7943 | -0.141 | -0.2161 | No |
| 47 | Abcd1 | 8077 | -0.150 | -0.2207 | No |
| 48 | Aldh9a1 | 8450 | -0.176 | -0.2435 | No |
| 49 | Hras | 8484 | -0.177 | -0.2388 | No |
| 50 | Pabpc1 | 8529 | -0.181 | -0.2348 | No |
| 51 | Elovl5 | 8675 | -0.192 | -0.2386 | No |
| 52 | Siah1a | 9312 | -0.240 | -0.2801 | No |
| 53 | Fads1 | 9770 | -0.279 | -0.3055 | Yes |
| 54 | Slc25a19 | 9798 | -0.283 | -0.2959 | Yes |
| 55 | Pex2 | 9914 | -0.293 | -0.2931 | Yes |
| 56 | Prdx5 | 9930 | -0.294 | -0.2821 | Yes |
| 57 | Top2a | 9959 | -0.297 | -0.2720 | Yes |
| 58 | Abcb9 | 10036 | -0.304 | -0.2655 | Yes |
| 59 | Ercc1 | 10135 | -0.311 | -0.2606 | Yes |
| 60 | Retsat | 10305 | -0.326 | -0.2607 | Yes |
| 61 | Crat | 10336 | -0.330 | -0.2494 | Yes |
| 62 | Slc25a4 | 10350 | -0.331 | -0.2368 | Yes |
| 63 | Vps4b | 10474 | -0.344 | -0.2324 | Yes |
| 64 | Acox1 | 10685 | -0.370 | -0.2340 | Yes |
| 65 | Abcb1a | 10728 | -0.376 | -0.2218 | Yes |
| 66 | Abcd3 | 10782 | -0.382 | -0.2103 | Yes |
| 67 | Hsd3b7 | 10846 | -0.390 | -0.1992 | Yes |
| 68 | Rdh11 | 10941 | -0.401 | -0.1901 | Yes |
| 69 | Fdps | 11077 | -0.419 | -0.1836 | Yes |
| 70 | Itgb1bp1 | 11116 | -0.425 | -0.1691 | Yes |
| 71 | Abcd2 | 11119 | -0.425 | -0.1516 | Yes |
| 72 | Hmgcl | 11385 | -0.471 | -0.1534 | Yes |
| 73 | Ehhadh | 11484 | -0.489 | -0.1411 | Yes |
| 74 | Acsl1 | 11508 | -0.494 | -0.1224 | Yes |
| 75 | Scp2 | 11522 | -0.498 | -0.1028 | Yes |
| 76 | Sod2 | 11907 | -0.595 | -0.1092 | Yes |
| 77 | Idh1 | 11973 | -0.617 | -0.0888 | Yes |
| 78 | Eci2 | 11974 | -0.617 | -0.0632 | Yes |
| 79 | Cdk7 | 12188 | -0.716 | -0.0507 | Yes |
| 80 | Prdx1 | 12266 | -0.779 | -0.0245 | Yes |
| 81 | Bcl10 | 12375 | -0.930 | 0.0053 | Yes |