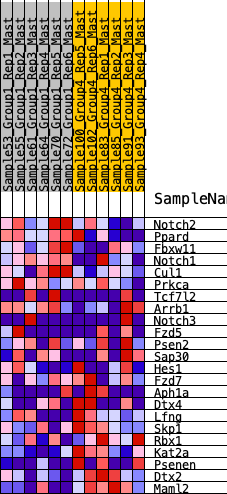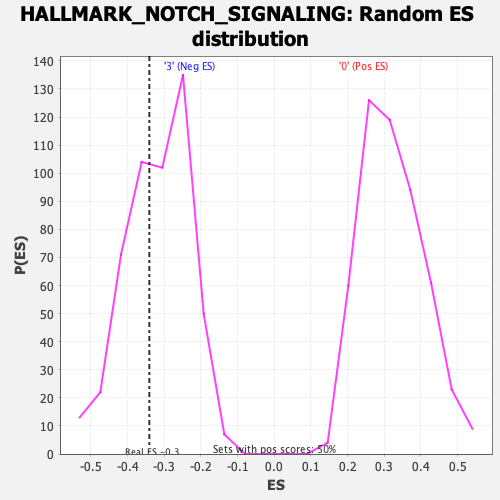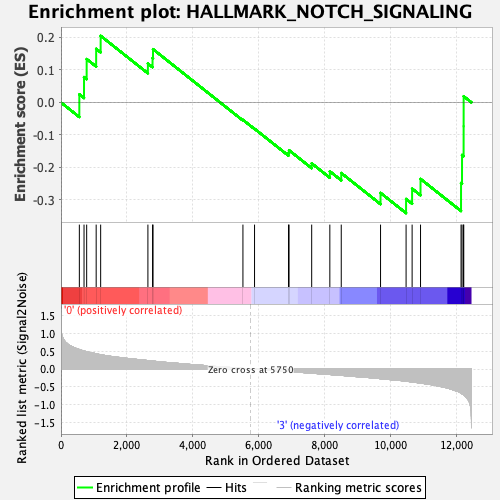
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | -0.34050092 |
| Normalized Enrichment Score (NES) | -1.0688714 |
| Nominal p-value | 0.37103173 |
| FDR q-value | 0.531204 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Notch2 | 556 | 0.549 | 0.0249 | No |
| 2 | Ppard | 698 | 0.507 | 0.0781 | No |
| 3 | Fbxw11 | 778 | 0.487 | 0.1336 | No |
| 4 | Notch1 | 1066 | 0.430 | 0.1651 | No |
| 5 | Cul1 | 1202 | 0.399 | 0.2049 | No |
| 6 | Prkca | 2634 | 0.237 | 0.1198 | No |
| 7 | Tcf7l2 | 2775 | 0.222 | 0.1368 | No |
| 8 | Arrb1 | 2789 | 0.222 | 0.1639 | No |
| 9 | Notch3 | 5514 | 0.016 | -0.0534 | No |
| 10 | Fzd5 | 5866 | -0.006 | -0.0810 | No |
| 11 | Psen2 | 6895 | -0.070 | -0.1548 | No |
| 12 | Sap30 | 6912 | -0.071 | -0.1471 | No |
| 13 | Hes1 | 7598 | -0.117 | -0.1873 | No |
| 14 | Fzd7 | 8148 | -0.154 | -0.2119 | No |
| 15 | Aph1a | 8494 | -0.178 | -0.2171 | No |
| 16 | Dtx4 | 9685 | -0.272 | -0.2783 | No |
| 17 | Lfng | 10459 | -0.343 | -0.2969 | Yes |
| 18 | Skp1 | 10642 | -0.365 | -0.2651 | Yes |
| 19 | Rbx1 | 10897 | -0.396 | -0.2353 | Yes |
| 20 | Kat2a | 12125 | -0.679 | -0.2478 | Yes |
| 21 | Psenen | 12155 | -0.697 | -0.1615 | Yes |
| 22 | Dtx2 | 12203 | -0.725 | -0.0732 | Yes |
| 23 | Maml2 | 12204 | -0.726 | 0.0191 | Yes |