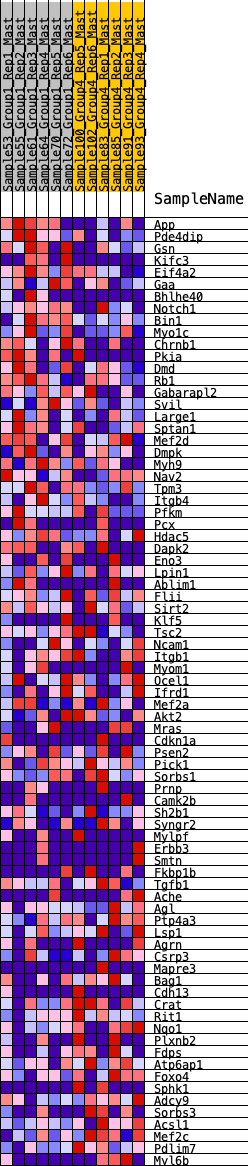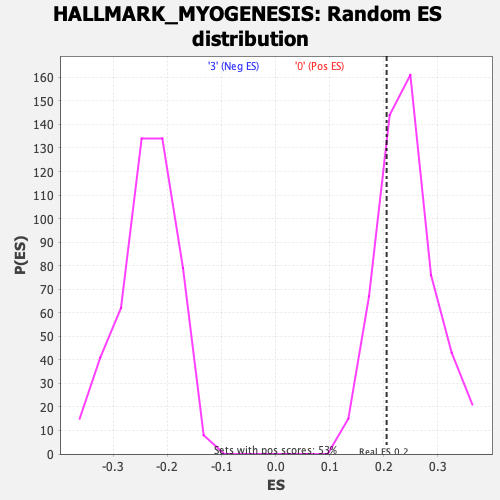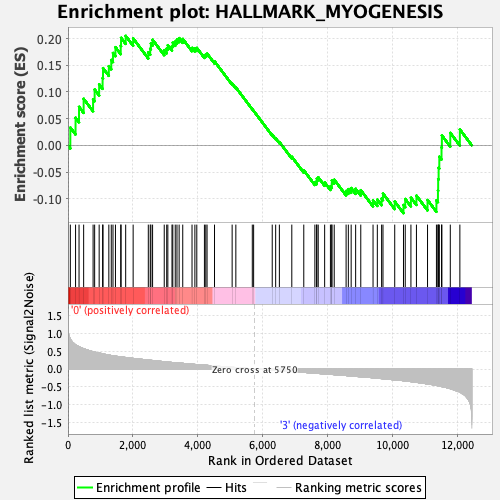
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.20514463 |
| Normalized Enrichment Score (NES) | 0.8496272 |
| Nominal p-value | 0.7628083 |
| FDR q-value | 0.90667766 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | App | 72 | 0.844 | 0.0333 | Yes |
| 2 | Pde4dip | 233 | 0.675 | 0.0517 | Yes |
| 3 | Gsn | 340 | 0.626 | 0.0722 | Yes |
| 4 | Kifc3 | 482 | 0.570 | 0.0872 | Yes |
| 5 | Eif4a2 | 773 | 0.488 | 0.0864 | Yes |
| 6 | Gaa | 821 | 0.474 | 0.1046 | Yes |
| 7 | Bhlhe40 | 960 | 0.451 | 0.1144 | Yes |
| 8 | Notch1 | 1066 | 0.430 | 0.1258 | Yes |
| 9 | Bin1 | 1079 | 0.427 | 0.1447 | Yes |
| 10 | Myo1c | 1258 | 0.391 | 0.1484 | Yes |
| 11 | Chrnb1 | 1333 | 0.380 | 0.1601 | Yes |
| 12 | Pkia | 1386 | 0.372 | 0.1731 | Yes |
| 13 | Dmd | 1462 | 0.363 | 0.1839 | Yes |
| 14 | Rb1 | 1625 | 0.339 | 0.1865 | Yes |
| 15 | Gabarapl2 | 1635 | 0.338 | 0.2015 | Yes |
| 16 | Svil | 1777 | 0.324 | 0.2051 | Yes |
| 17 | Large1 | 2007 | 0.298 | 0.2004 | No |
| 18 | Sptan1 | 2474 | 0.251 | 0.1744 | No |
| 19 | Mef2d | 2533 | 0.247 | 0.1812 | No |
| 20 | Dmpk | 2554 | 0.244 | 0.1909 | No |
| 21 | Myh9 | 2606 | 0.239 | 0.1979 | No |
| 22 | Nav2 | 2965 | 0.203 | 0.1783 | No |
| 23 | Tpm3 | 3041 | 0.197 | 0.1814 | No |
| 24 | Itgb4 | 3076 | 0.194 | 0.1876 | No |
| 25 | Pfkm | 3204 | 0.184 | 0.1859 | No |
| 26 | Pcx | 3229 | 0.183 | 0.1924 | No |
| 27 | Hdac5 | 3304 | 0.178 | 0.1947 | No |
| 28 | Dapk2 | 3357 | 0.173 | 0.1985 | No |
| 29 | Eno3 | 3424 | 0.167 | 0.2009 | No |
| 30 | Lpin1 | 3535 | 0.159 | 0.1994 | No |
| 31 | Ablim1 | 3821 | 0.136 | 0.1827 | No |
| 32 | Flii | 3909 | 0.128 | 0.1816 | No |
| 33 | Sirt2 | 3968 | 0.124 | 0.1827 | No |
| 34 | Klf5 | 4203 | 0.109 | 0.1688 | No |
| 35 | Tsc2 | 4245 | 0.106 | 0.1704 | No |
| 36 | Ncam1 | 4284 | 0.103 | 0.1721 | No |
| 37 | Itgb1 | 4514 | 0.085 | 0.1575 | No |
| 38 | Myom1 | 5057 | 0.048 | 0.1158 | No |
| 39 | Ocel1 | 5171 | 0.039 | 0.1085 | No |
| 40 | Ifrd1 | 5678 | 0.004 | 0.0678 | No |
| 41 | Mef2a | 5719 | 0.002 | 0.0646 | No |
| 42 | Akt2 | 6292 | -0.033 | 0.0199 | No |
| 43 | Mras | 6398 | -0.040 | 0.0132 | No |
| 44 | Cdkn1a | 6512 | -0.047 | 0.0063 | No |
| 45 | Psen2 | 6895 | -0.070 | -0.0214 | No |
| 46 | Pick1 | 7264 | -0.095 | -0.0467 | No |
| 47 | Sorbs1 | 7606 | -0.118 | -0.0688 | No |
| 48 | Prnp | 7659 | -0.121 | -0.0674 | No |
| 49 | Camk2b | 7667 | -0.122 | -0.0623 | No |
| 50 | Sh2b1 | 7709 | -0.125 | -0.0599 | No |
| 51 | Syngr2 | 7907 | -0.138 | -0.0694 | No |
| 52 | Mylpf | 8085 | -0.151 | -0.0767 | No |
| 53 | Erbb3 | 8128 | -0.153 | -0.0730 | No |
| 54 | Smtn | 8129 | -0.153 | -0.0659 | No |
| 55 | Fkbp1b | 8202 | -0.159 | -0.0643 | No |
| 56 | Tgfb1 | 8568 | -0.184 | -0.0853 | No |
| 57 | Ache | 8637 | -0.189 | -0.0821 | No |
| 58 | Agl | 8724 | -0.196 | -0.0799 | No |
| 59 | Ptp4a3 | 8862 | -0.208 | -0.0814 | No |
| 60 | Lsp1 | 9020 | -0.217 | -0.0840 | No |
| 61 | Agrn | 9398 | -0.247 | -0.1030 | No |
| 62 | Csrp3 | 9530 | -0.259 | -0.1016 | No |
| 63 | Mapre3 | 9662 | -0.270 | -0.0996 | No |
| 64 | Bag1 | 9704 | -0.273 | -0.0903 | No |
| 65 | Cdh13 | 10066 | -0.306 | -0.1052 | No |
| 66 | Crat | 10336 | -0.330 | -0.1117 | No |
| 67 | Rit1 | 10390 | -0.336 | -0.1004 | No |
| 68 | Nqo1 | 10565 | -0.355 | -0.0980 | No |
| 69 | Plxnb2 | 10735 | -0.376 | -0.0942 | No |
| 70 | Fdps | 11077 | -0.419 | -0.1024 | No |
| 71 | Atp6ap1 | 11351 | -0.464 | -0.1029 | No |
| 72 | Foxo4 | 11398 | -0.474 | -0.0847 | No |
| 73 | Sphk1 | 11405 | -0.476 | -0.0631 | No |
| 74 | Adcy9 | 11419 | -0.478 | -0.0419 | No |
| 75 | Sorbs3 | 11440 | -0.482 | -0.0212 | No |
| 76 | Acsl1 | 11508 | -0.494 | -0.0037 | No |
| 77 | Mef2c | 11519 | -0.497 | 0.0185 | No |
| 78 | Pdlim7 | 11778 | -0.554 | 0.0234 | No |
| 79 | Myl6b | 12073 | -0.651 | 0.0298 | No |