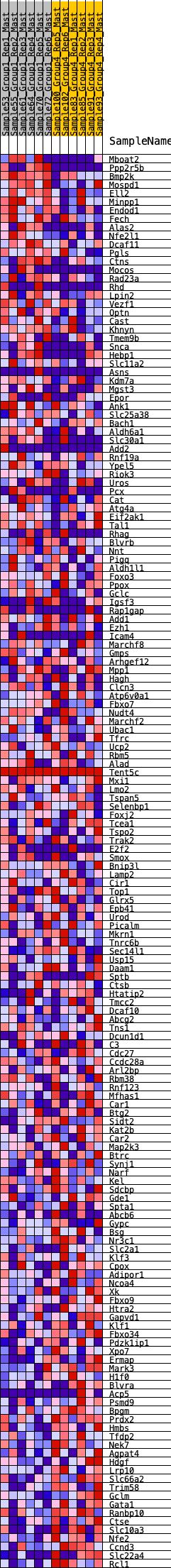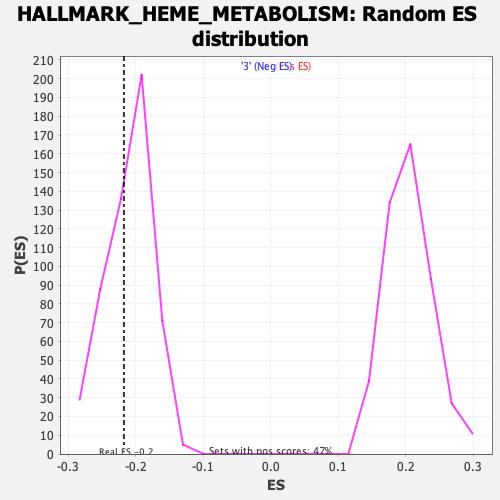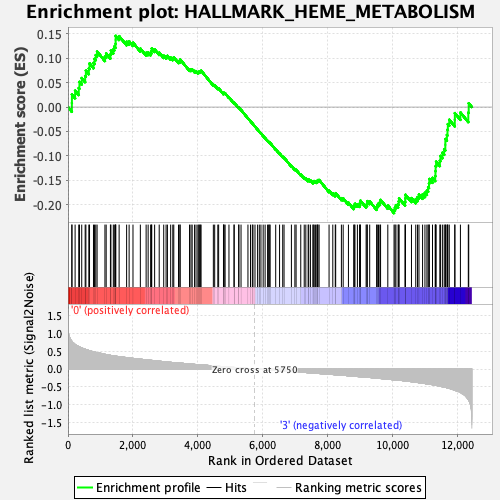
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_HEME_METABOLISM |
| Enrichment Score (ES) | -0.21721767 |
| Normalized Enrichment Score (NES) | -1.0369571 |
| Nominal p-value | 0.39548022 |
| FDR q-value | 0.54658645 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mboat2 | 119 | 0.781 | 0.0082 | No |
| 2 | Ppp2r5b | 121 | 0.776 | 0.0258 | No |
| 3 | Bmp2k | 219 | 0.688 | 0.0337 | No |
| 4 | Mospd1 | 332 | 0.630 | 0.0390 | No |
| 5 | Ell2 | 353 | 0.622 | 0.0516 | No |
| 6 | Minpp1 | 423 | 0.592 | 0.0595 | No |
| 7 | Endod1 | 529 | 0.557 | 0.0636 | No |
| 8 | Fech | 551 | 0.550 | 0.0745 | No |
| 9 | Alas2 | 642 | 0.520 | 0.0791 | No |
| 10 | Nfe2l1 | 663 | 0.516 | 0.0893 | No |
| 11 | Dcaf11 | 785 | 0.482 | 0.0904 | No |
| 12 | Pgls | 818 | 0.475 | 0.0987 | No |
| 13 | Ctns | 856 | 0.468 | 0.1064 | No |
| 14 | Mocos | 896 | 0.460 | 0.1137 | No |
| 15 | Rad23a | 1138 | 0.413 | 0.1035 | No |
| 16 | Rhd | 1176 | 0.406 | 0.1098 | No |
| 17 | Lpin2 | 1307 | 0.384 | 0.1080 | No |
| 18 | Vezf1 | 1322 | 0.381 | 0.1156 | No |
| 19 | Optn | 1396 | 0.370 | 0.1181 | No |
| 20 | Cast | 1427 | 0.368 | 0.1241 | No |
| 21 | Khnyn | 1464 | 0.363 | 0.1295 | No |
| 22 | Tmem9b | 1468 | 0.362 | 0.1375 | No |
| 23 | Snca | 1469 | 0.361 | 0.1457 | No |
| 24 | Hebp1 | 1575 | 0.347 | 0.1451 | No |
| 25 | Slc11a2 | 1806 | 0.321 | 0.1337 | No |
| 26 | Asns | 1883 | 0.311 | 0.1346 | No |
| 27 | Kdm7a | 2002 | 0.299 | 0.1319 | No |
| 28 | Mgst3 | 2229 | 0.275 | 0.1197 | No |
| 29 | Epor | 2407 | 0.256 | 0.1112 | No |
| 30 | Ank1 | 2469 | 0.251 | 0.1120 | No |
| 31 | Slc25a38 | 2546 | 0.245 | 0.1114 | No |
| 32 | Bach1 | 2572 | 0.243 | 0.1149 | No |
| 33 | Aldh6a1 | 2580 | 0.242 | 0.1198 | No |
| 34 | Slc30a1 | 2669 | 0.234 | 0.1180 | No |
| 35 | Add2 | 2810 | 0.220 | 0.1116 | No |
| 36 | Rnf19a | 2949 | 0.205 | 0.1051 | No |
| 37 | Ypel5 | 3022 | 0.199 | 0.1038 | No |
| 38 | Riok3 | 3063 | 0.195 | 0.1050 | No |
| 39 | Uros | 3163 | 0.186 | 0.1012 | No |
| 40 | Pcx | 3229 | 0.183 | 0.1001 | No |
| 41 | Cat | 3259 | 0.180 | 0.1018 | No |
| 42 | Atg4a | 3405 | 0.168 | 0.0939 | No |
| 43 | Eif2ak1 | 3447 | 0.165 | 0.0943 | No |
| 44 | Tal1 | 3455 | 0.164 | 0.0975 | No |
| 45 | Rhag | 3744 | 0.142 | 0.0772 | No |
| 46 | Blvrb | 3788 | 0.139 | 0.0769 | No |
| 47 | Nnt | 3825 | 0.136 | 0.0771 | No |
| 48 | Pigq | 3899 | 0.130 | 0.0741 | No |
| 49 | Aldh1l1 | 3945 | 0.126 | 0.0733 | No |
| 50 | Foxo3 | 4001 | 0.122 | 0.0716 | No |
| 51 | Ppox | 4028 | 0.120 | 0.0722 | No |
| 52 | Gclc | 4056 | 0.117 | 0.0727 | No |
| 53 | Igsf3 | 4078 | 0.116 | 0.0737 | No |
| 54 | Rap1gap | 4100 | 0.115 | 0.0746 | No |
| 55 | Add1 | 4478 | 0.087 | 0.0459 | No |
| 56 | Ezh1 | 4519 | 0.085 | 0.0445 | No |
| 57 | Icam4 | 4616 | 0.078 | 0.0385 | No |
| 58 | Marchf8 | 4641 | 0.076 | 0.0383 | No |
| 59 | Gmps | 4790 | 0.065 | 0.0277 | No |
| 60 | Arhgef12 | 4791 | 0.065 | 0.0292 | No |
| 61 | Mpp1 | 4812 | 0.063 | 0.0290 | No |
| 62 | Hagh | 4840 | 0.061 | 0.0282 | No |
| 63 | Clcn3 | 4959 | 0.054 | 0.0198 | No |
| 64 | Atp6v0a1 | 5116 | 0.044 | 0.0081 | No |
| 65 | Fbxo7 | 5119 | 0.044 | 0.0090 | No |
| 66 | Nudt4 | 5259 | 0.033 | -0.0016 | No |
| 67 | Marchf2 | 5265 | 0.033 | -0.0013 | No |
| 68 | Ubac1 | 5331 | 0.028 | -0.0059 | No |
| 69 | Tfrc | 5544 | 0.014 | -0.0229 | No |
| 70 | Ucp2 | 5623 | 0.009 | -0.0290 | No |
| 71 | Rbm5 | 5679 | 0.004 | -0.0334 | No |
| 72 | Alad | 5699 | 0.003 | -0.0349 | No |
| 73 | Tent5c | 5761 | 0.000 | -0.0399 | No |
| 74 | Mxi1 | 5841 | -0.004 | -0.0462 | No |
| 75 | Lmo2 | 5894 | -0.008 | -0.0503 | No |
| 76 | Tspan5 | 5944 | -0.011 | -0.0540 | No |
| 77 | Selenbp1 | 6013 | -0.015 | -0.0592 | No |
| 78 | Foxj2 | 6073 | -0.019 | -0.0636 | No |
| 79 | Tcea1 | 6149 | -0.023 | -0.0691 | No |
| 80 | Tspo2 | 6182 | -0.025 | -0.0712 | No |
| 81 | Trak2 | 6202 | -0.026 | -0.0721 | No |
| 82 | E2f2 | 6228 | -0.028 | -0.0735 | No |
| 83 | Smox | 6399 | -0.040 | -0.0865 | No |
| 84 | Bnip3l | 6518 | -0.048 | -0.0950 | No |
| 85 | Lamp2 | 6615 | -0.053 | -0.1016 | No |
| 86 | Cir1 | 6656 | -0.055 | -0.1036 | No |
| 87 | Top1 | 6883 | -0.069 | -0.1204 | No |
| 88 | Glrx5 | 6990 | -0.077 | -0.1273 | No |
| 89 | Epb41 | 7027 | -0.078 | -0.1284 | No |
| 90 | Urod | 7171 | -0.088 | -0.1381 | No |
| 91 | Picalm | 7278 | -0.096 | -0.1445 | No |
| 92 | Mkrn1 | 7327 | -0.098 | -0.1462 | No |
| 93 | Tnrc6b | 7399 | -0.104 | -0.1496 | No |
| 94 | Sec14l1 | 7416 | -0.105 | -0.1485 | No |
| 95 | Usp15 | 7469 | -0.109 | -0.1502 | No |
| 96 | Daam1 | 7548 | -0.114 | -0.1540 | No |
| 97 | Sptb | 7554 | -0.114 | -0.1518 | No |
| 98 | Ctsb | 7586 | -0.116 | -0.1516 | No |
| 99 | Htatip2 | 7628 | -0.119 | -0.1522 | No |
| 100 | Tmcc2 | 7668 | -0.122 | -0.1526 | No |
| 101 | Dcaf10 | 7685 | -0.123 | -0.1511 | No |
| 102 | Abcg2 | 7700 | -0.124 | -0.1494 | No |
| 103 | Tns1 | 7736 | -0.126 | -0.1494 | No |
| 104 | Dcun1d1 | 8043 | -0.148 | -0.1709 | No |
| 105 | C3 | 8158 | -0.155 | -0.1767 | No |
| 106 | Cdc27 | 8234 | -0.161 | -0.1791 | No |
| 107 | Ccdc28a | 8247 | -0.162 | -0.1764 | No |
| 108 | Arl2bp | 8423 | -0.174 | -0.1866 | No |
| 109 | Rbm38 | 8478 | -0.177 | -0.1870 | No |
| 110 | Rnf123 | 8640 | -0.189 | -0.1958 | No |
| 111 | Mfhas1 | 8803 | -0.203 | -0.2044 | No |
| 112 | Car1 | 8821 | -0.204 | -0.2011 | No |
| 113 | Btg2 | 8841 | -0.206 | -0.1979 | No |
| 114 | Sidt2 | 8916 | -0.211 | -0.1991 | No |
| 115 | Kat2b | 8983 | -0.215 | -0.1996 | No |
| 116 | Car2 | 8999 | -0.216 | -0.1958 | No |
| 117 | Map2k3 | 9009 | -0.217 | -0.1916 | No |
| 118 | Btrc | 9185 | -0.230 | -0.2006 | No |
| 119 | Synj1 | 9213 | -0.232 | -0.1975 | No |
| 120 | Narf | 9219 | -0.232 | -0.1926 | No |
| 121 | Kel | 9295 | -0.238 | -0.1933 | No |
| 122 | Sdcbp | 9508 | -0.258 | -0.2047 | No |
| 123 | Gde1 | 9528 | -0.259 | -0.2003 | No |
| 124 | Spta1 | 9566 | -0.262 | -0.1973 | No |
| 125 | Abcb6 | 9610 | -0.266 | -0.1947 | No |
| 126 | Gypc | 9630 | -0.268 | -0.1901 | No |
| 127 | Bsg | 9853 | -0.287 | -0.2017 | No |
| 128 | Nr3c1 | 10045 | -0.305 | -0.2102 | Yes |
| 129 | Slc2a1 | 10074 | -0.306 | -0.2055 | Yes |
| 130 | Klf3 | 10103 | -0.308 | -0.2008 | Yes |
| 131 | Cpox | 10170 | -0.313 | -0.1990 | Yes |
| 132 | Adipor1 | 10188 | -0.315 | -0.1931 | Yes |
| 133 | Ncoa4 | 10200 | -0.316 | -0.1868 | Yes |
| 134 | Xk | 10385 | -0.335 | -0.1941 | Yes |
| 135 | Fbxo9 | 10392 | -0.336 | -0.1869 | Yes |
| 136 | Htra2 | 10397 | -0.336 | -0.1796 | Yes |
| 137 | Gapvd1 | 10582 | -0.357 | -0.1864 | Yes |
| 138 | Klf1 | 10714 | -0.374 | -0.1885 | Yes |
| 139 | Fbxo34 | 10769 | -0.380 | -0.1842 | Yes |
| 140 | Pdzk1ip1 | 10813 | -0.386 | -0.1789 | Yes |
| 141 | Xpo7 | 10924 | -0.399 | -0.1788 | Yes |
| 142 | Ermap | 10997 | -0.409 | -0.1753 | Yes |
| 143 | Mark3 | 11058 | -0.417 | -0.1706 | Yes |
| 144 | H1f0 | 11096 | -0.423 | -0.1640 | Yes |
| 145 | Blvra | 11122 | -0.425 | -0.1563 | Yes |
| 146 | Acp5 | 11132 | -0.427 | -0.1472 | Yes |
| 147 | Psmd9 | 11230 | -0.443 | -0.1450 | Yes |
| 148 | Bpgm | 11313 | -0.458 | -0.1412 | Yes |
| 149 | Prdx2 | 11321 | -0.460 | -0.1313 | Yes |
| 150 | Hmbs | 11326 | -0.461 | -0.1211 | Yes |
| 151 | Tfdp2 | 11341 | -0.463 | -0.1116 | Yes |
| 152 | Nek7 | 11457 | -0.484 | -0.1099 | Yes |
| 153 | Agpat4 | 11474 | -0.487 | -0.1001 | Yes |
| 154 | Hdgf | 11534 | -0.500 | -0.0934 | Yes |
| 155 | Lrp10 | 11590 | -0.511 | -0.0862 | Yes |
| 156 | Slc66a2 | 11625 | -0.517 | -0.0772 | Yes |
| 157 | Trim58 | 11626 | -0.517 | -0.0654 | Yes |
| 158 | Gclm | 11674 | -0.528 | -0.0571 | Yes |
| 159 | Gata1 | 11692 | -0.532 | -0.0463 | Yes |
| 160 | Ranbp10 | 11703 | -0.534 | -0.0349 | Yes |
| 161 | Ctse | 11746 | -0.544 | -0.0259 | Yes |
| 162 | Slc10a3 | 11917 | -0.600 | -0.0261 | Yes |
| 163 | Nfe2 | 11919 | -0.600 | -0.0124 | Yes |
| 164 | Ccnd3 | 12090 | -0.661 | -0.0112 | Yes |
| 165 | Slc22a4 | 12333 | -0.856 | -0.0113 | Yes |
| 166 | Rcl1 | 12346 | -0.875 | 0.0077 | Yes |