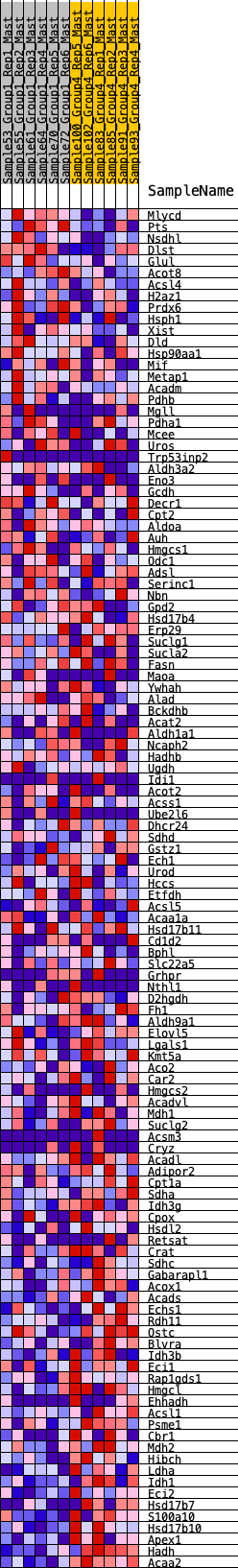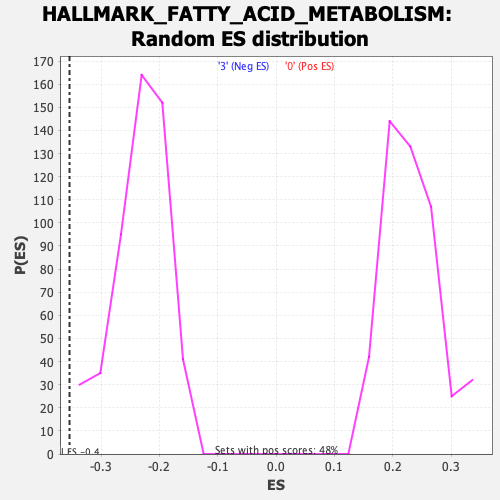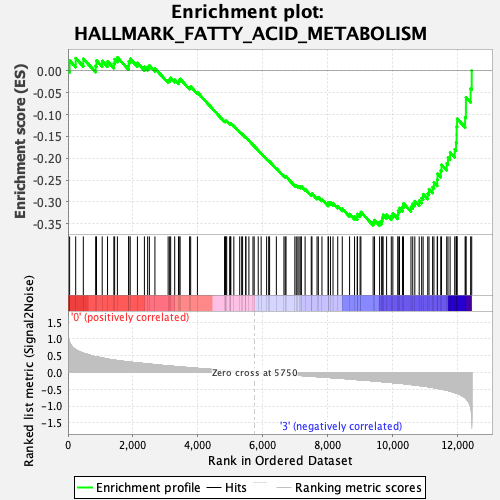
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.35408312 |
| Normalized Enrichment Score (NES) | -1.526579 |
| Nominal p-value | 0.003868472 |
| FDR q-value | 0.14958097 |
| FWER p-Value | 0.225 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mlycd | 50 | 0.890 | 0.0233 | No |
| 2 | Pts | 237 | 0.674 | 0.0289 | No |
| 3 | Nsdhl | 472 | 0.574 | 0.0276 | No |
| 4 | Dlst | 852 | 0.469 | 0.0112 | No |
| 5 | Glul | 878 | 0.463 | 0.0234 | No |
| 6 | Acot8 | 1054 | 0.431 | 0.0225 | No |
| 7 | Acsl4 | 1218 | 0.397 | 0.0215 | No |
| 8 | H2az1 | 1413 | 0.369 | 0.0171 | No |
| 9 | Prdx6 | 1437 | 0.367 | 0.0265 | No |
| 10 | Hsph1 | 1522 | 0.354 | 0.0305 | No |
| 11 | Xist | 1868 | 0.313 | 0.0122 | No |
| 12 | Dld | 1873 | 0.313 | 0.0214 | No |
| 13 | Hsp90aa1 | 1917 | 0.307 | 0.0274 | No |
| 14 | Mif | 2139 | 0.284 | 0.0182 | No |
| 15 | Metap1 | 2354 | 0.263 | 0.0089 | No |
| 16 | Acadm | 2454 | 0.252 | 0.0086 | No |
| 17 | Pdhb | 2507 | 0.249 | 0.0121 | No |
| 18 | Mgll | 2678 | 0.233 | 0.0054 | No |
| 19 | Pdha1 | 3085 | 0.193 | -0.0216 | No |
| 20 | Mcee | 3132 | 0.189 | -0.0195 | No |
| 21 | Uros | 3163 | 0.186 | -0.0162 | No |
| 22 | Trp53inp2 | 3287 | 0.178 | -0.0207 | No |
| 23 | Aldh3a2 | 3404 | 0.168 | -0.0249 | No |
| 24 | Eno3 | 3424 | 0.167 | -0.0214 | No |
| 25 | Gcdh | 3454 | 0.164 | -0.0187 | No |
| 26 | Decr1 | 3746 | 0.142 | -0.0379 | No |
| 27 | Cpt2 | 3779 | 0.140 | -0.0362 | No |
| 28 | Aldoa | 3987 | 0.122 | -0.0493 | No |
| 29 | Auh | 4823 | 0.063 | -0.1151 | No |
| 30 | Hmgcs1 | 4851 | 0.061 | -0.1154 | No |
| 31 | Odc1 | 4864 | 0.059 | -0.1146 | No |
| 32 | Adsl | 4889 | 0.058 | -0.1147 | No |
| 33 | Serinc1 | 4981 | 0.052 | -0.1205 | No |
| 34 | Nbn | 5008 | 0.050 | -0.1211 | No |
| 35 | Gpd2 | 5016 | 0.050 | -0.1201 | No |
| 36 | Hsd17b4 | 5109 | 0.044 | -0.1262 | No |
| 37 | Erp29 | 5290 | 0.031 | -0.1399 | No |
| 38 | Suclg1 | 5351 | 0.026 | -0.1440 | No |
| 39 | Sucla2 | 5379 | 0.025 | -0.1454 | No |
| 40 | Fasn | 5476 | 0.019 | -0.1526 | No |
| 41 | Maoa | 5480 | 0.019 | -0.1523 | No |
| 42 | Ywhah | 5573 | 0.012 | -0.1594 | No |
| 43 | Alad | 5699 | 0.003 | -0.1694 | No |
| 44 | Bckdhb | 5736 | 0.001 | -0.1723 | No |
| 45 | Acat2 | 5861 | -0.005 | -0.1822 | No |
| 46 | Aldh1a1 | 5951 | -0.012 | -0.1891 | No |
| 47 | Ncaph2 | 6119 | -0.021 | -0.2020 | No |
| 48 | Hadhb | 6183 | -0.025 | -0.2063 | No |
| 49 | Ugdh | 6211 | -0.027 | -0.2077 | No |
| 50 | Idi1 | 6419 | -0.041 | -0.2232 | No |
| 51 | Acot2 | 6653 | -0.055 | -0.2404 | No |
| 52 | Acss1 | 6692 | -0.057 | -0.2417 | No |
| 53 | Ube2l6 | 6717 | -0.059 | -0.2419 | No |
| 54 | Dhcr24 | 6989 | -0.077 | -0.2615 | No |
| 55 | Sdhd | 7033 | -0.079 | -0.2626 | No |
| 56 | Gstz1 | 7082 | -0.082 | -0.2640 | No |
| 57 | Ech1 | 7120 | -0.084 | -0.2644 | No |
| 58 | Urod | 7171 | -0.088 | -0.2657 | No |
| 59 | Hccs | 7190 | -0.089 | -0.2645 | No |
| 60 | Etfdh | 7305 | -0.097 | -0.2707 | No |
| 61 | Acsl5 | 7494 | -0.110 | -0.2826 | No |
| 62 | Acaa1a | 7516 | -0.112 | -0.2808 | No |
| 63 | Hsd17b11 | 7673 | -0.122 | -0.2897 | No |
| 64 | Cd1d2 | 7713 | -0.125 | -0.2891 | No |
| 65 | Bphl | 7824 | -0.132 | -0.2939 | No |
| 66 | Slc22a5 | 8016 | -0.146 | -0.3049 | No |
| 67 | Grhpr | 8019 | -0.146 | -0.3006 | No |
| 68 | Nthl1 | 8086 | -0.151 | -0.3013 | No |
| 69 | D2hgdh | 8165 | -0.156 | -0.3029 | No |
| 70 | Fh1 | 8310 | -0.166 | -0.3094 | No |
| 71 | Aldh9a1 | 8450 | -0.176 | -0.3153 | No |
| 72 | Elovl5 | 8675 | -0.192 | -0.3276 | No |
| 73 | Lgals1 | 8828 | -0.205 | -0.3337 | No |
| 74 | Kmt5a | 8913 | -0.211 | -0.3340 | No |
| 75 | Aco2 | 8915 | -0.211 | -0.3276 | No |
| 76 | Car2 | 8999 | -0.216 | -0.3277 | No |
| 77 | Hmgcs2 | 9024 | -0.218 | -0.3229 | No |
| 78 | Acadvl | 9401 | -0.247 | -0.3458 | No |
| 79 | Mdh1 | 9444 | -0.252 | -0.3415 | No |
| 80 | Suclg2 | 9600 | -0.265 | -0.3459 | Yes |
| 81 | Acsm3 | 9664 | -0.270 | -0.3427 | Yes |
| 82 | Cryz | 9682 | -0.272 | -0.3358 | Yes |
| 83 | Acadl | 9708 | -0.274 | -0.3294 | Yes |
| 84 | Adipor2 | 9816 | -0.284 | -0.3293 | Yes |
| 85 | Cpt1a | 9960 | -0.297 | -0.3318 | Yes |
| 86 | Sdha | 10008 | -0.301 | -0.3264 | Yes |
| 87 | Idh3g | 10163 | -0.313 | -0.3292 | Yes |
| 88 | Cpox | 10170 | -0.313 | -0.3201 | Yes |
| 89 | Hsdl2 | 10214 | -0.318 | -0.3138 | Yes |
| 90 | Retsat | 10305 | -0.326 | -0.3111 | Yes |
| 91 | Crat | 10336 | -0.330 | -0.3034 | Yes |
| 92 | Sdhc | 10567 | -0.355 | -0.3111 | Yes |
| 93 | Gabarapl1 | 10621 | -0.363 | -0.3043 | Yes |
| 94 | Acox1 | 10685 | -0.370 | -0.2980 | Yes |
| 95 | Acads | 10820 | -0.387 | -0.2970 | Yes |
| 96 | Echs1 | 10896 | -0.396 | -0.2909 | Yes |
| 97 | Rdh11 | 10941 | -0.401 | -0.2822 | Yes |
| 98 | Ostc | 11083 | -0.420 | -0.2807 | Yes |
| 99 | Blvra | 11122 | -0.425 | -0.2707 | Yes |
| 100 | Idh3b | 11233 | -0.444 | -0.2660 | Yes |
| 101 | Eci1 | 11274 | -0.451 | -0.2554 | Yes |
| 102 | Rap1gds1 | 11375 | -0.468 | -0.2491 | Yes |
| 103 | Hmgcl | 11385 | -0.471 | -0.2354 | Yes |
| 104 | Ehhadh | 11484 | -0.489 | -0.2283 | Yes |
| 105 | Acsl1 | 11508 | -0.494 | -0.2150 | Yes |
| 106 | Psme1 | 11668 | -0.526 | -0.2117 | Yes |
| 107 | Cbr1 | 11707 | -0.535 | -0.1984 | Yes |
| 108 | Mdh2 | 11772 | -0.552 | -0.1866 | Yes |
| 109 | Hibch | 11915 | -0.598 | -0.1797 | Yes |
| 110 | Ldha | 11956 | -0.611 | -0.1642 | Yes |
| 111 | Idh1 | 11973 | -0.617 | -0.1465 | Yes |
| 112 | Eci2 | 11974 | -0.617 | -0.1276 | Yes |
| 113 | Hsd17b7 | 11989 | -0.623 | -0.1096 | Yes |
| 114 | S100a10 | 12235 | -0.746 | -0.1065 | Yes |
| 115 | Hsd17b10 | 12259 | -0.776 | -0.0845 | Yes |
| 116 | Apex1 | 12260 | -0.776 | -0.0607 | Yes |
| 117 | Hadh | 12405 | -1.025 | -0.0409 | Yes |
| 118 | Acaa2 | 12438 | -1.422 | 0.0002 | Yes |