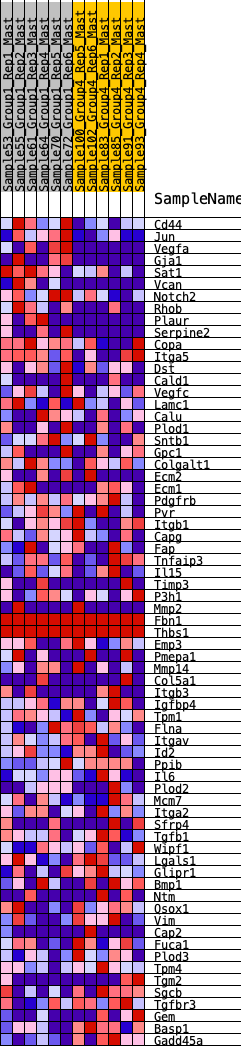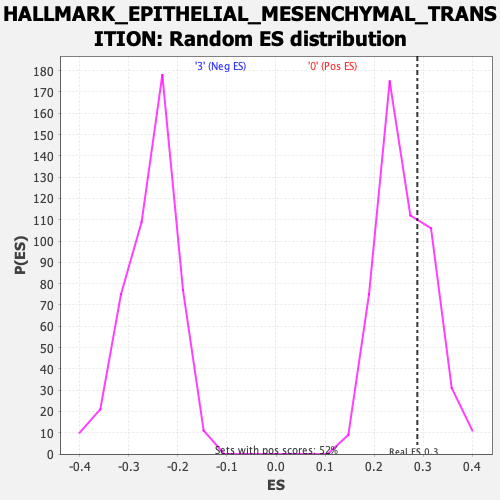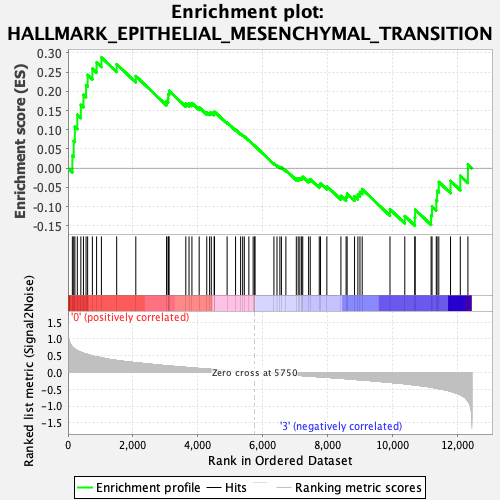
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2884158 |
| Normalized Enrichment Score (NES) | 1.1003172 |
| Nominal p-value | 0.33333334 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd44 | 134 | 0.761 | 0.0327 | Yes |
| 2 | Jun | 176 | 0.726 | 0.0709 | Yes |
| 3 | Vegfa | 212 | 0.695 | 0.1078 | Yes |
| 4 | Gja1 | 287 | 0.653 | 0.1392 | Yes |
| 5 | Sat1 | 396 | 0.601 | 0.1648 | Yes |
| 6 | Vcan | 476 | 0.573 | 0.1912 | Yes |
| 7 | Notch2 | 556 | 0.549 | 0.2162 | Yes |
| 8 | Rhob | 605 | 0.535 | 0.2429 | Yes |
| 9 | Plaur | 751 | 0.493 | 0.2593 | Yes |
| 10 | Serpine2 | 881 | 0.463 | 0.2754 | Yes |
| 11 | Copa | 1029 | 0.436 | 0.2884 | Yes |
| 12 | Itga5 | 1500 | 0.357 | 0.2708 | No |
| 13 | Dst | 2089 | 0.289 | 0.2398 | No |
| 14 | Cald1 | 3036 | 0.198 | 0.1747 | No |
| 15 | Vegfc | 3086 | 0.193 | 0.1818 | No |
| 16 | Lamc1 | 3091 | 0.193 | 0.1925 | No |
| 17 | Calu | 3115 | 0.190 | 0.2015 | No |
| 18 | Plod1 | 3629 | 0.151 | 0.1687 | No |
| 19 | Sntb1 | 3729 | 0.144 | 0.1689 | No |
| 20 | Gpc1 | 3818 | 0.136 | 0.1696 | No |
| 21 | Colgalt1 | 4041 | 0.118 | 0.1584 | No |
| 22 | Ecm2 | 4275 | 0.103 | 0.1455 | No |
| 23 | Ecm1 | 4363 | 0.096 | 0.1440 | No |
| 24 | Pdgfrb | 4415 | 0.092 | 0.1451 | No |
| 25 | Pvr | 4502 | 0.086 | 0.1430 | No |
| 26 | Itgb1 | 4514 | 0.085 | 0.1470 | No |
| 27 | Capg | 4902 | 0.057 | 0.1190 | No |
| 28 | Fap | 5162 | 0.040 | 0.1003 | No |
| 29 | Tnfaip3 | 5318 | 0.028 | 0.0894 | No |
| 30 | Il15 | 5381 | 0.025 | 0.0858 | No |
| 31 | Timp3 | 5432 | 0.021 | 0.0829 | No |
| 32 | P3h1 | 5572 | 0.012 | 0.0724 | No |
| 33 | Mmp2 | 5704 | 0.002 | 0.0619 | No |
| 34 | Fbn1 | 5753 | 0.000 | 0.0580 | No |
| 35 | Thbs1 | 5759 | 0.000 | 0.0576 | No |
| 36 | Emp3 | 6344 | -0.036 | 0.0125 | No |
| 37 | Pmepa1 | 6439 | -0.043 | 0.0074 | No |
| 38 | Mmp14 | 6528 | -0.048 | 0.0030 | No |
| 39 | Col5a1 | 6575 | -0.051 | 0.0022 | No |
| 40 | Itgb3 | 6713 | -0.058 | -0.0056 | No |
| 41 | Igfbp4 | 7034 | -0.079 | -0.0269 | No |
| 42 | Tpm1 | 7092 | -0.082 | -0.0268 | No |
| 43 | Flna | 7143 | -0.086 | -0.0259 | No |
| 44 | Itgav | 7198 | -0.090 | -0.0251 | No |
| 45 | Id2 | 7232 | -0.093 | -0.0225 | No |
| 46 | Ppib | 7413 | -0.105 | -0.0310 | No |
| 47 | Il6 | 7463 | -0.108 | -0.0288 | No |
| 48 | Plod2 | 7741 | -0.127 | -0.0439 | No |
| 49 | Mcm7 | 7780 | -0.130 | -0.0396 | No |
| 50 | Itga2 | 7975 | -0.143 | -0.0471 | No |
| 51 | Sfrp4 | 8408 | -0.172 | -0.0721 | No |
| 52 | Tgfb1 | 8568 | -0.184 | -0.0745 | No |
| 53 | Wipf1 | 8599 | -0.186 | -0.0663 | No |
| 54 | Lgals1 | 8828 | -0.205 | -0.0730 | No |
| 55 | Glipr1 | 8931 | -0.213 | -0.0691 | No |
| 56 | Bmp1 | 8995 | -0.216 | -0.0618 | No |
| 57 | Ntm | 9064 | -0.220 | -0.0547 | No |
| 58 | Qsox1 | 9920 | -0.293 | -0.1070 | No |
| 59 | Vim | 10376 | -0.334 | -0.1247 | No |
| 60 | Cap2 | 10684 | -0.370 | -0.1284 | No |
| 61 | Fuca1 | 10695 | -0.371 | -0.1080 | No |
| 62 | Plod3 | 11187 | -0.434 | -0.1228 | No |
| 63 | Tpm4 | 11215 | -0.440 | -0.0998 | No |
| 64 | Tgm2 | 11346 | -0.464 | -0.0838 | No |
| 65 | Sgcb | 11371 | -0.467 | -0.0591 | No |
| 66 | Tgfbr3 | 11424 | -0.480 | -0.0358 | No |
| 67 | Gem | 11785 | -0.555 | -0.0332 | No |
| 68 | Basp1 | 12086 | -0.659 | -0.0197 | No |
| 69 | Gadd45a | 12321 | -0.845 | 0.0097 | No |