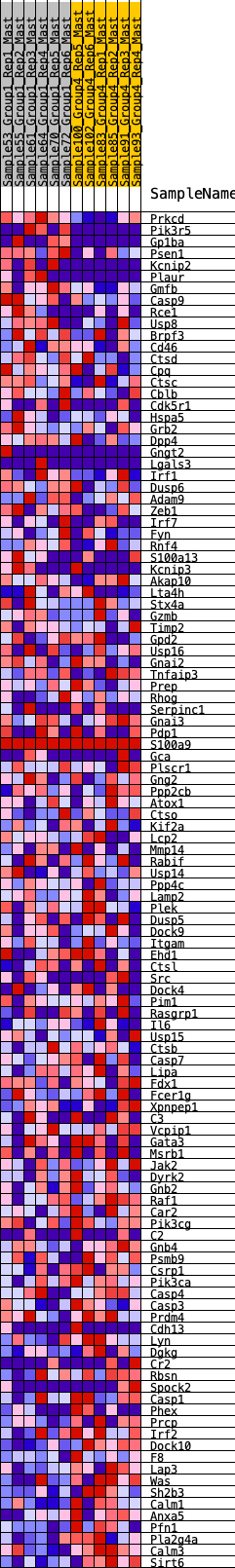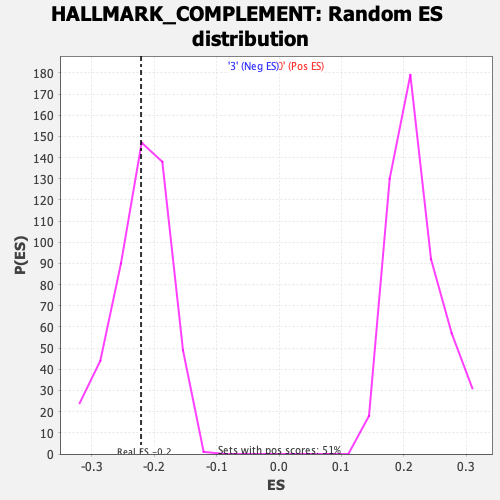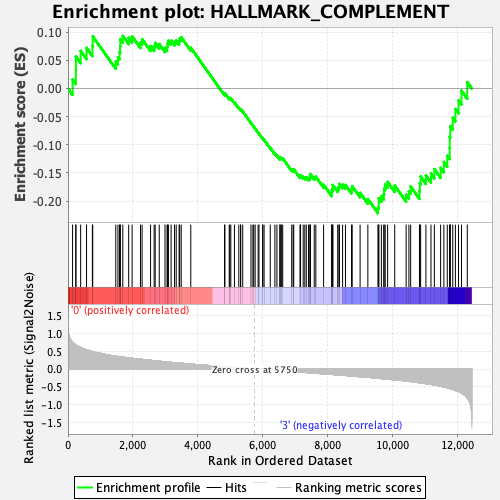
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.22129808 |
| Normalized Enrichment Score (NES) | -0.9988381 |
| Nominal p-value | 0.43204868 |
| FDR q-value | 0.6015 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkcd | 141 | 0.752 | 0.0159 | No |
| 2 | Pik3r5 | 241 | 0.672 | 0.0323 | No |
| 3 | Gp1ba | 243 | 0.672 | 0.0567 | No |
| 4 | Psen1 | 390 | 0.603 | 0.0668 | No |
| 5 | Kcnip2 | 571 | 0.545 | 0.0720 | No |
| 6 | Plaur | 751 | 0.493 | 0.0754 | No |
| 7 | Gmfb | 762 | 0.491 | 0.0924 | No |
| 8 | Casp9 | 1472 | 0.361 | 0.0480 | No |
| 9 | Rce1 | 1539 | 0.351 | 0.0555 | No |
| 10 | Usp8 | 1588 | 0.346 | 0.0642 | No |
| 11 | Brpf3 | 1605 | 0.343 | 0.0753 | No |
| 12 | Cd46 | 1612 | 0.342 | 0.0873 | No |
| 13 | Ctsd | 1688 | 0.333 | 0.0933 | No |
| 14 | Cpq | 1870 | 0.313 | 0.0900 | No |
| 15 | Ctsc | 1975 | 0.301 | 0.0925 | No |
| 16 | Cblb | 2233 | 0.274 | 0.0817 | No |
| 17 | Cdk5r1 | 2283 | 0.271 | 0.0875 | No |
| 18 | Hspa5 | 2544 | 0.245 | 0.0754 | No |
| 19 | Grb2 | 2654 | 0.234 | 0.0750 | No |
| 20 | Dpp4 | 2686 | 0.232 | 0.0810 | No |
| 21 | Gngt2 | 2811 | 0.220 | 0.0789 | No |
| 22 | Lgals3 | 2989 | 0.201 | 0.0718 | No |
| 23 | Irf1 | 3052 | 0.196 | 0.0739 | No |
| 24 | Dusp6 | 3067 | 0.195 | 0.0799 | No |
| 25 | Adam9 | 3094 | 0.192 | 0.0848 | No |
| 26 | Zeb1 | 3176 | 0.186 | 0.0850 | No |
| 27 | Irf7 | 3283 | 0.179 | 0.0829 | No |
| 28 | Fyn | 3334 | 0.176 | 0.0852 | No |
| 29 | Rnf4 | 3423 | 0.167 | 0.0841 | No |
| 30 | S100a13 | 3441 | 0.165 | 0.0887 | No |
| 31 | Kcnip3 | 3488 | 0.163 | 0.0909 | No |
| 32 | Akap10 | 3783 | 0.139 | 0.0721 | No |
| 33 | Lta4h | 4822 | 0.063 | -0.0098 | No |
| 34 | Stx4a | 4834 | 0.062 | -0.0084 | No |
| 35 | Gzmb | 4967 | 0.053 | -0.0172 | No |
| 36 | Timp2 | 4985 | 0.052 | -0.0167 | No |
| 37 | Gpd2 | 5016 | 0.050 | -0.0173 | No |
| 38 | Usp16 | 5128 | 0.043 | -0.0248 | No |
| 39 | Gnai2 | 5262 | 0.033 | -0.0344 | No |
| 40 | Tnfaip3 | 5318 | 0.028 | -0.0378 | No |
| 41 | Prep | 5319 | 0.028 | -0.0368 | No |
| 42 | Rhog | 5383 | 0.024 | -0.0410 | No |
| 43 | Serpinc1 | 5637 | 0.008 | -0.0612 | No |
| 44 | Gnai3 | 5696 | 0.003 | -0.0658 | No |
| 45 | Pdp1 | 5723 | 0.001 | -0.0679 | No |
| 46 | S100a9 | 5766 | 0.000 | -0.0713 | No |
| 47 | Gca | 5857 | -0.005 | -0.0784 | No |
| 48 | Plscr1 | 5884 | -0.007 | -0.0803 | No |
| 49 | Gng2 | 5997 | -0.014 | -0.0889 | No |
| 50 | Ppp2cb | 6000 | -0.014 | -0.0885 | No |
| 51 | Atox1 | 6041 | -0.016 | -0.0912 | No |
| 52 | Ctso | 6231 | -0.028 | -0.1055 | No |
| 53 | Kif2a | 6374 | -0.038 | -0.1156 | No |
| 54 | Lcp2 | 6436 | -0.042 | -0.1190 | No |
| 55 | Mmp14 | 6528 | -0.048 | -0.1246 | No |
| 56 | Rabif | 6532 | -0.049 | -0.1231 | No |
| 57 | Usp14 | 6558 | -0.050 | -0.1233 | No |
| 58 | Ppp4c | 6583 | -0.051 | -0.1234 | No |
| 59 | Lamp2 | 6615 | -0.053 | -0.1240 | No |
| 60 | Plek | 6893 | -0.070 | -0.1439 | No |
| 61 | Dusp5 | 6936 | -0.073 | -0.1447 | No |
| 62 | Dock9 | 6955 | -0.074 | -0.1434 | No |
| 63 | Itgam | 7148 | -0.086 | -0.1558 | No |
| 64 | Ehd1 | 7164 | -0.087 | -0.1539 | No |
| 65 | Ctsl | 7246 | -0.094 | -0.1570 | No |
| 66 | Src | 7296 | -0.096 | -0.1575 | No |
| 67 | Dock4 | 7341 | -0.100 | -0.1575 | No |
| 68 | Pim1 | 7409 | -0.104 | -0.1591 | No |
| 69 | Rasgrp1 | 7450 | -0.107 | -0.1584 | No |
| 70 | Il6 | 7463 | -0.108 | -0.1555 | No |
| 71 | Usp15 | 7469 | -0.109 | -0.1519 | No |
| 72 | Ctsb | 7586 | -0.116 | -0.1571 | No |
| 73 | Casp7 | 7635 | -0.120 | -0.1567 | No |
| 74 | Lipa | 7875 | -0.137 | -0.1711 | No |
| 75 | Fdx1 | 8123 | -0.153 | -0.1856 | No |
| 76 | Fcer1g | 8126 | -0.153 | -0.1801 | No |
| 77 | Xpnpep1 | 8156 | -0.155 | -0.1769 | No |
| 78 | C3 | 8158 | -0.155 | -0.1713 | No |
| 79 | Vcpip1 | 8309 | -0.166 | -0.1774 | No |
| 80 | Gata3 | 8347 | -0.168 | -0.1743 | No |
| 81 | Msrb1 | 8363 | -0.170 | -0.1694 | No |
| 82 | Jak2 | 8460 | -0.176 | -0.1707 | No |
| 83 | Dyrk2 | 8548 | -0.182 | -0.1712 | No |
| 84 | Gnb2 | 8735 | -0.196 | -0.1791 | No |
| 85 | Raf1 | 8756 | -0.198 | -0.1735 | No |
| 86 | Car2 | 8999 | -0.216 | -0.1853 | No |
| 87 | Pik3cg | 9238 | -0.233 | -0.1961 | No |
| 88 | C2 | 9549 | -0.261 | -0.2118 | Yes |
| 89 | Gnb4 | 9573 | -0.263 | -0.2041 | Yes |
| 90 | Psmb9 | 9574 | -0.263 | -0.1946 | Yes |
| 91 | Csrp1 | 9655 | -0.270 | -0.1913 | Yes |
| 92 | Pik3ca | 9729 | -0.276 | -0.1872 | Yes |
| 93 | Casp4 | 9739 | -0.277 | -0.1778 | Yes |
| 94 | Casp3 | 9773 | -0.279 | -0.1703 | Yes |
| 95 | Prdm4 | 9847 | -0.286 | -0.1658 | Yes |
| 96 | Cdh13 | 10066 | -0.306 | -0.1724 | Yes |
| 97 | Lyn | 10418 | -0.338 | -0.1886 | Yes |
| 98 | Dgkg | 10503 | -0.348 | -0.1827 | Yes |
| 99 | Cr2 | 10557 | -0.354 | -0.1742 | Yes |
| 100 | Rbsn | 10828 | -0.388 | -0.1820 | Yes |
| 101 | Spock2 | 10838 | -0.389 | -0.1686 | Yes |
| 102 | Casp1 | 10864 | -0.392 | -0.1563 | Yes |
| 103 | Phex | 11027 | -0.414 | -0.1544 | Yes |
| 104 | Prcp | 11183 | -0.433 | -0.1512 | Yes |
| 105 | Irf2 | 11287 | -0.454 | -0.1431 | Yes |
| 106 | Dock10 | 11480 | -0.488 | -0.1409 | Yes |
| 107 | F8 | 11580 | -0.509 | -0.1304 | Yes |
| 108 | Lap3 | 11685 | -0.530 | -0.1196 | Yes |
| 109 | Was | 11758 | -0.546 | -0.1055 | Yes |
| 110 | Sh2b3 | 11761 | -0.548 | -0.0858 | Yes |
| 111 | Calm1 | 11779 | -0.554 | -0.0670 | Yes |
| 112 | Anxa5 | 11854 | -0.577 | -0.0520 | Yes |
| 113 | Pfn1 | 11935 | -0.606 | -0.0365 | Yes |
| 114 | Pla2g4a | 12032 | -0.633 | -0.0212 | Yes |
| 115 | Calm3 | 12123 | -0.675 | -0.0040 | Yes |
| 116 | Sirt6 | 12301 | -0.817 | 0.0114 | Yes |