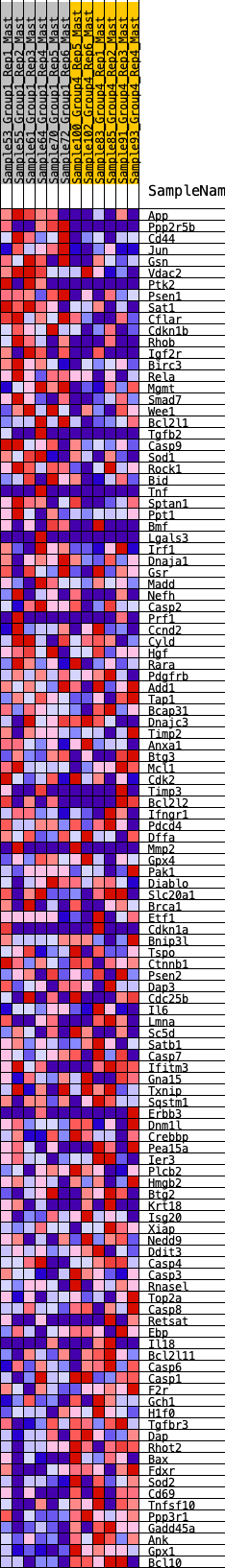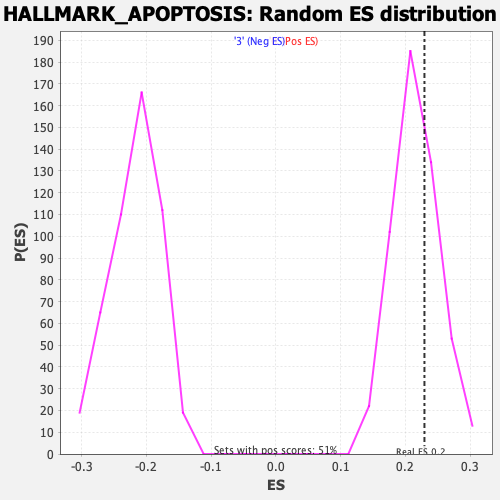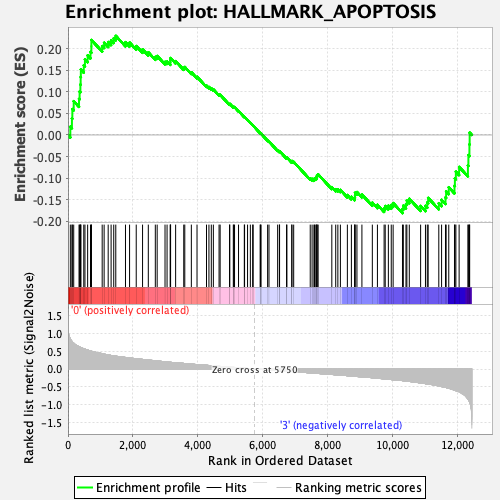
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.22929195 |
| Normalized Enrichment Score (NES) | 1.0584472 |
| Nominal p-value | 0.31827113 |
| FDR q-value | 0.8394622 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | App | 72 | 0.844 | 0.0192 | Yes |
| 2 | Ppp2r5b | 121 | 0.776 | 0.0383 | Yes |
| 3 | Cd44 | 134 | 0.761 | 0.0600 | Yes |
| 4 | Jun | 176 | 0.726 | 0.0782 | Yes |
| 5 | Gsn | 340 | 0.626 | 0.0835 | Yes |
| 6 | Vdac2 | 358 | 0.620 | 0.1006 | Yes |
| 7 | Ptk2 | 382 | 0.608 | 0.1167 | Yes |
| 8 | Psen1 | 390 | 0.603 | 0.1341 | Yes |
| 9 | Sat1 | 396 | 0.601 | 0.1515 | Yes |
| 10 | Cflar | 486 | 0.569 | 0.1612 | Yes |
| 11 | Cdkn1b | 522 | 0.558 | 0.1749 | Yes |
| 12 | Rhob | 605 | 0.535 | 0.1841 | Yes |
| 13 | Igf2r | 693 | 0.509 | 0.1922 | Yes |
| 14 | Birc3 | 720 | 0.501 | 0.2049 | Yes |
| 15 | Rela | 721 | 0.500 | 0.2198 | Yes |
| 16 | Mgmt | 1053 | 0.431 | 0.2057 | Yes |
| 17 | Smad7 | 1113 | 0.420 | 0.2134 | Yes |
| 18 | Wee1 | 1238 | 0.394 | 0.2151 | Yes |
| 19 | Bcl2l1 | 1328 | 0.381 | 0.2191 | Yes |
| 20 | Tgfb2 | 1409 | 0.370 | 0.2236 | Yes |
| 21 | Casp9 | 1472 | 0.361 | 0.2293 | Yes |
| 22 | Sod1 | 1771 | 0.325 | 0.2147 | No |
| 23 | Rock1 | 1897 | 0.309 | 0.2138 | No |
| 24 | Bid | 2102 | 0.287 | 0.2058 | No |
| 25 | Tnf | 2297 | 0.269 | 0.1980 | No |
| 26 | Sptan1 | 2474 | 0.251 | 0.1912 | No |
| 27 | Ppt1 | 2690 | 0.232 | 0.1806 | No |
| 28 | Bmf | 2747 | 0.225 | 0.1828 | No |
| 29 | Lgals3 | 2989 | 0.201 | 0.1692 | No |
| 30 | Irf1 | 3052 | 0.196 | 0.1700 | No |
| 31 | Dnaja1 | 3151 | 0.188 | 0.1676 | No |
| 32 | Gsr | 3153 | 0.187 | 0.1731 | No |
| 33 | Madd | 3161 | 0.186 | 0.1780 | No |
| 34 | Nefh | 3316 | 0.178 | 0.1708 | No |
| 35 | Casp2 | 3566 | 0.156 | 0.1552 | No |
| 36 | Prf1 | 3598 | 0.154 | 0.1573 | No |
| 37 | Ccnd2 | 3800 | 0.138 | 0.1451 | No |
| 38 | Cyld | 3975 | 0.123 | 0.1346 | No |
| 39 | Hgf | 4267 | 0.104 | 0.1141 | No |
| 40 | Rara | 4342 | 0.098 | 0.1110 | No |
| 41 | Pdgfrb | 4415 | 0.092 | 0.1079 | No |
| 42 | Add1 | 4478 | 0.087 | 0.1054 | No |
| 43 | Tap1 | 4654 | 0.075 | 0.0935 | No |
| 44 | Bcap31 | 4687 | 0.072 | 0.0930 | No |
| 45 | Dnajc3 | 4976 | 0.052 | 0.0712 | No |
| 46 | Timp2 | 4985 | 0.052 | 0.0720 | No |
| 47 | Anxa1 | 5093 | 0.045 | 0.0647 | No |
| 48 | Btg3 | 5106 | 0.045 | 0.0651 | No |
| 49 | Mcl1 | 5131 | 0.042 | 0.0644 | No |
| 50 | Cdk2 | 5254 | 0.034 | 0.0555 | No |
| 51 | Timp3 | 5432 | 0.021 | 0.0417 | No |
| 52 | Bcl2l2 | 5440 | 0.021 | 0.0418 | No |
| 53 | Ifngr1 | 5532 | 0.015 | 0.0348 | No |
| 54 | Pdcd4 | 5618 | 0.009 | 0.0282 | No |
| 55 | Dffa | 5690 | 0.003 | 0.0225 | No |
| 56 | Mmp2 | 5704 | 0.002 | 0.0215 | No |
| 57 | Gpx4 | 5921 | -0.010 | 0.0043 | No |
| 58 | Pak1 | 5929 | -0.010 | 0.0040 | No |
| 59 | Diablo | 5948 | -0.011 | 0.0029 | No |
| 60 | Slc20a1 | 6150 | -0.024 | -0.0127 | No |
| 61 | Brca1 | 6192 | -0.026 | -0.0152 | No |
| 62 | Etf1 | 6459 | -0.044 | -0.0355 | No |
| 63 | Cdkn1a | 6512 | -0.047 | -0.0383 | No |
| 64 | Bnip3l | 6518 | -0.048 | -0.0373 | No |
| 65 | Tspo | 6733 | -0.060 | -0.0529 | No |
| 66 | Ctnnb1 | 6744 | -0.060 | -0.0519 | No |
| 67 | Psen2 | 6895 | -0.070 | -0.0620 | No |
| 68 | Dap3 | 6898 | -0.070 | -0.0601 | No |
| 69 | Cdc25b | 6953 | -0.074 | -0.0623 | No |
| 70 | Il6 | 7463 | -0.108 | -0.1004 | No |
| 71 | Lmna | 7520 | -0.112 | -0.1016 | No |
| 72 | Sc5d | 7577 | -0.116 | -0.1027 | No |
| 73 | Satb1 | 7594 | -0.117 | -0.1005 | No |
| 74 | Casp7 | 7635 | -0.120 | -0.1002 | No |
| 75 | Ifitm3 | 7660 | -0.121 | -0.0986 | No |
| 76 | Gna15 | 7664 | -0.122 | -0.0952 | No |
| 77 | Txnip | 7678 | -0.123 | -0.0926 | No |
| 78 | Sqstm1 | 7706 | -0.125 | -0.0911 | No |
| 79 | Erbb3 | 8128 | -0.153 | -0.1207 | No |
| 80 | Dnm1l | 8248 | -0.162 | -0.1255 | No |
| 81 | Crebbp | 8314 | -0.166 | -0.1259 | No |
| 82 | Pea15a | 8392 | -0.171 | -0.1270 | No |
| 83 | Ier3 | 8607 | -0.187 | -0.1389 | No |
| 84 | Plcb2 | 8731 | -0.196 | -0.1430 | No |
| 85 | Hmgb2 | 8831 | -0.205 | -0.1450 | No |
| 86 | Btg2 | 8841 | -0.206 | -0.1396 | No |
| 87 | Krt18 | 8843 | -0.206 | -0.1336 | No |
| 88 | Isg20 | 8902 | -0.210 | -0.1320 | No |
| 89 | Xiap | 9055 | -0.220 | -0.1379 | No |
| 90 | Nedd9 | 9376 | -0.246 | -0.1565 | No |
| 91 | Ddit3 | 9533 | -0.260 | -0.1615 | No |
| 92 | Casp4 | 9739 | -0.277 | -0.1699 | No |
| 93 | Casp3 | 9773 | -0.279 | -0.1643 | No |
| 94 | Rnasel | 9870 | -0.288 | -0.1635 | No |
| 95 | Top2a | 9959 | -0.297 | -0.1619 | No |
| 96 | Casp8 | 10017 | -0.301 | -0.1575 | No |
| 97 | Retsat | 10305 | -0.326 | -0.1711 | No |
| 98 | Ebp | 10331 | -0.329 | -0.1634 | No |
| 99 | Il18 | 10413 | -0.337 | -0.1600 | No |
| 100 | Bcl2l11 | 10440 | -0.341 | -0.1520 | No |
| 101 | Casp6 | 10515 | -0.349 | -0.1476 | No |
| 102 | Casp1 | 10864 | -0.392 | -0.1642 | No |
| 103 | F2r | 11013 | -0.412 | -0.1640 | No |
| 104 | Gch1 | 11069 | -0.419 | -0.1560 | No |
| 105 | H1f0 | 11096 | -0.423 | -0.1456 | No |
| 106 | Tgfbr3 | 11424 | -0.480 | -0.1579 | No |
| 107 | Dap | 11510 | -0.494 | -0.1501 | No |
| 108 | Rhot2 | 11630 | -0.518 | -0.1444 | No |
| 109 | Bax | 11650 | -0.522 | -0.1305 | No |
| 110 | Fdxr | 11731 | -0.540 | -0.1209 | No |
| 111 | Sod2 | 11907 | -0.595 | -0.1175 | No |
| 112 | Cd69 | 11920 | -0.600 | -0.1006 | No |
| 113 | Tnfsf10 | 11950 | -0.610 | -0.0849 | No |
| 114 | Ppp3r1 | 12050 | -0.640 | -0.0739 | No |
| 115 | Gadd45a | 12321 | -0.845 | -0.0707 | No |
| 116 | Ank | 12336 | -0.856 | -0.0465 | No |
| 117 | Gpx1 | 12368 | -0.917 | -0.0218 | No |
| 118 | Bcl10 | 12375 | -0.930 | 0.0054 | No |