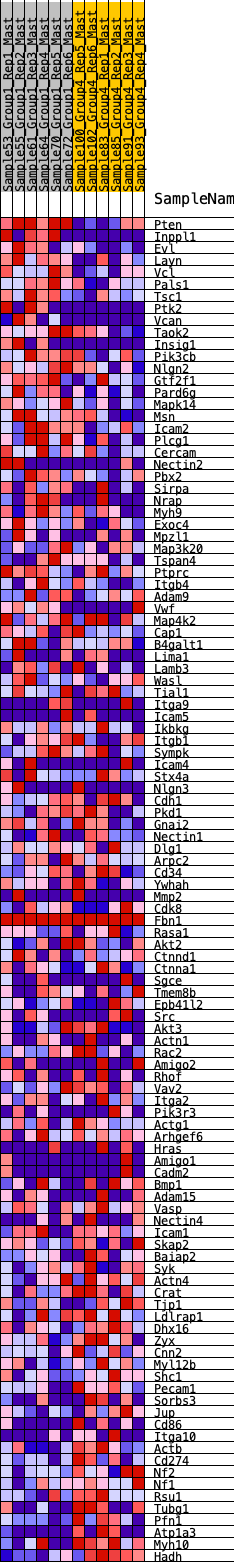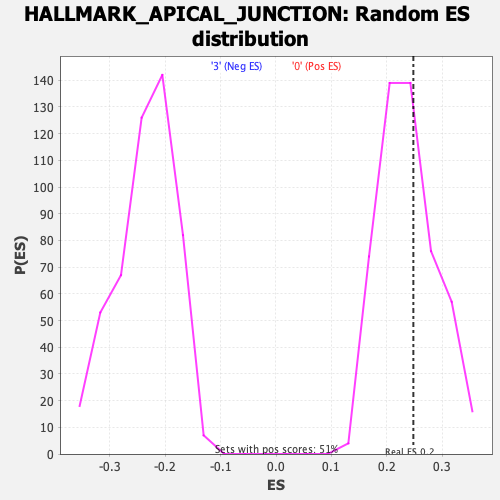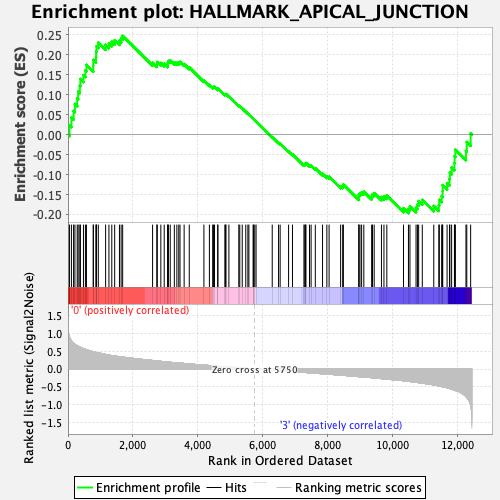
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.24760853 |
| Normalized Enrichment Score (NES) | 1.0379311 |
| Nominal p-value | 0.37623763 |
| FDR q-value | 0.7519304 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pten | 45 | 0.905 | 0.0236 | Yes |
| 2 | Inppl1 | 105 | 0.794 | 0.0427 | Yes |
| 3 | Evl | 172 | 0.730 | 0.0593 | Yes |
| 4 | Layn | 213 | 0.695 | 0.0770 | Yes |
| 5 | Vcl | 284 | 0.654 | 0.0910 | Yes |
| 6 | Pals1 | 313 | 0.639 | 0.1080 | Yes |
| 7 | Tsc1 | 364 | 0.617 | 0.1225 | Yes |
| 8 | Ptk2 | 382 | 0.608 | 0.1394 | Yes |
| 9 | Vcan | 476 | 0.573 | 0.1491 | Yes |
| 10 | Taok2 | 536 | 0.554 | 0.1610 | Yes |
| 11 | Insig1 | 568 | 0.545 | 0.1749 | Yes |
| 12 | Pik3cb | 779 | 0.486 | 0.1725 | Yes |
| 13 | Nlgn2 | 780 | 0.486 | 0.1871 | Yes |
| 14 | Gtf2f1 | 863 | 0.467 | 0.1946 | Yes |
| 15 | Pard6g | 867 | 0.466 | 0.2084 | Yes |
| 16 | Mapk14 | 879 | 0.463 | 0.2214 | Yes |
| 17 | Msn | 934 | 0.456 | 0.2307 | Yes |
| 18 | Icam2 | 1159 | 0.409 | 0.2249 | Yes |
| 19 | Plcg1 | 1261 | 0.391 | 0.2285 | Yes |
| 20 | Cercam | 1349 | 0.378 | 0.2328 | Yes |
| 21 | Nectin2 | 1438 | 0.366 | 0.2367 | Yes |
| 22 | Pbx2 | 1585 | 0.346 | 0.2353 | Yes |
| 23 | Sirpa | 1637 | 0.338 | 0.2413 | Yes |
| 24 | Nrap | 1684 | 0.334 | 0.2476 | Yes |
| 25 | Myh9 | 2606 | 0.239 | 0.1801 | No |
| 26 | Exoc4 | 2736 | 0.227 | 0.1765 | No |
| 27 | Mpzl1 | 2750 | 0.225 | 0.1822 | No |
| 28 | Map3k20 | 2860 | 0.214 | 0.1798 | No |
| 29 | Tspan4 | 2964 | 0.203 | 0.1776 | No |
| 30 | Ptprc | 3068 | 0.195 | 0.1751 | No |
| 31 | Itgb4 | 3076 | 0.194 | 0.1804 | No |
| 32 | Adam9 | 3094 | 0.192 | 0.1848 | No |
| 33 | Vwf | 3150 | 0.188 | 0.1860 | No |
| 34 | Map4k2 | 3276 | 0.179 | 0.1812 | No |
| 35 | Cap1 | 3354 | 0.173 | 0.1802 | No |
| 36 | B4galt1 | 3413 | 0.168 | 0.1805 | No |
| 37 | Lima1 | 3451 | 0.164 | 0.1825 | No |
| 38 | Lamb3 | 3578 | 0.155 | 0.1769 | No |
| 39 | Wasl | 3736 | 0.143 | 0.1685 | No |
| 40 | Tial1 | 4187 | 0.110 | 0.1353 | No |
| 41 | Itga9 | 4356 | 0.097 | 0.1246 | No |
| 42 | Icam5 | 4457 | 0.089 | 0.1192 | No |
| 43 | Ikbkg | 4485 | 0.087 | 0.1196 | No |
| 44 | Itgb1 | 4514 | 0.085 | 0.1199 | No |
| 45 | Sympk | 4615 | 0.078 | 0.1141 | No |
| 46 | Icam4 | 4616 | 0.078 | 0.1164 | No |
| 47 | Stx4a | 4834 | 0.062 | 0.1007 | No |
| 48 | Nlgn3 | 4849 | 0.061 | 0.1014 | No |
| 49 | Cdh1 | 4868 | 0.059 | 0.1017 | No |
| 50 | Pkd1 | 4957 | 0.054 | 0.0962 | No |
| 51 | Gnai2 | 5262 | 0.033 | 0.0725 | No |
| 52 | Nectin1 | 5292 | 0.031 | 0.0711 | No |
| 53 | Dlg1 | 5368 | 0.026 | 0.0658 | No |
| 54 | Arpc2 | 5477 | 0.019 | 0.0576 | No |
| 55 | Cd34 | 5542 | 0.014 | 0.0528 | No |
| 56 | Ywhah | 5573 | 0.012 | 0.0507 | No |
| 57 | Mmp2 | 5704 | 0.002 | 0.0403 | No |
| 58 | Cdk8 | 5713 | 0.002 | 0.0397 | No |
| 59 | Fbn1 | 5753 | 0.000 | 0.0365 | No |
| 60 | Rasa1 | 5794 | -0.001 | 0.0333 | No |
| 61 | Akt2 | 6292 | -0.033 | -0.0060 | No |
| 62 | Ctnnd1 | 6487 | -0.045 | -0.0204 | No |
| 63 | Ctnna1 | 6536 | -0.049 | -0.0228 | No |
| 64 | Sgce | 6796 | -0.064 | -0.0419 | No |
| 65 | Tmem8b | 6916 | -0.072 | -0.0494 | No |
| 66 | Epb41l2 | 7269 | -0.095 | -0.0751 | No |
| 67 | Src | 7296 | -0.096 | -0.0743 | No |
| 68 | Akt3 | 7314 | -0.097 | -0.0727 | No |
| 69 | Actn1 | 7331 | -0.099 | -0.0711 | No |
| 70 | Rac2 | 7441 | -0.107 | -0.0767 | No |
| 71 | Amigo2 | 7490 | -0.110 | -0.0773 | No |
| 72 | Rhof | 7619 | -0.119 | -0.0841 | No |
| 73 | Vav2 | 7842 | -0.134 | -0.0980 | No |
| 74 | Itga2 | 7975 | -0.143 | -0.1044 | No |
| 75 | Pik3r3 | 8044 | -0.148 | -0.1055 | No |
| 76 | Actg1 | 8399 | -0.172 | -0.1290 | No |
| 77 | Arhgef6 | 8463 | -0.176 | -0.1288 | No |
| 78 | Hras | 8484 | -0.177 | -0.1251 | No |
| 79 | Amigo1 | 8960 | -0.214 | -0.1572 | No |
| 80 | Cadm2 | 8961 | -0.214 | -0.1508 | No |
| 81 | Bmp1 | 8995 | -0.216 | -0.1469 | No |
| 82 | Adam15 | 9046 | -0.219 | -0.1444 | No |
| 83 | Vasp | 9114 | -0.224 | -0.1431 | No |
| 84 | Nectin4 | 9355 | -0.243 | -0.1552 | No |
| 85 | Icam1 | 9381 | -0.246 | -0.1498 | No |
| 86 | Skap2 | 9433 | -0.251 | -0.1464 | No |
| 87 | Baiap2 | 9657 | -0.270 | -0.1564 | No |
| 88 | Syk | 9734 | -0.276 | -0.1542 | No |
| 89 | Actn4 | 9823 | -0.284 | -0.1528 | No |
| 90 | Crat | 10336 | -0.330 | -0.1844 | No |
| 91 | Tjp1 | 10489 | -0.346 | -0.1863 | No |
| 92 | Ldlrap1 | 10535 | -0.352 | -0.1794 | No |
| 93 | Dhx16 | 10723 | -0.375 | -0.1833 | No |
| 94 | Zyx | 10766 | -0.380 | -0.1752 | No |
| 95 | Cnn2 | 10803 | -0.384 | -0.1666 | No |
| 96 | Myl12b | 10915 | -0.398 | -0.1636 | No |
| 97 | Shc1 | 11269 | -0.450 | -0.1787 | No |
| 98 | Pecam1 | 11425 | -0.480 | -0.1768 | No |
| 99 | Sorbs3 | 11440 | -0.482 | -0.1635 | No |
| 100 | Jup | 11512 | -0.494 | -0.1543 | No |
| 101 | Cd86 | 11541 | -0.501 | -0.1415 | No |
| 102 | Itga10 | 11543 | -0.501 | -0.1265 | No |
| 103 | Actb | 11679 | -0.529 | -0.1215 | No |
| 104 | Cd274 | 11753 | -0.546 | -0.1110 | No |
| 105 | Nf2 | 11765 | -0.549 | -0.0954 | No |
| 106 | Nf1 | 11819 | -0.567 | -0.0826 | No |
| 107 | Rsu1 | 11906 | -0.595 | -0.0717 | No |
| 108 | Tubg1 | 11911 | -0.597 | -0.0540 | No |
| 109 | Pfn1 | 11935 | -0.606 | -0.0376 | No |
| 110 | Atp1a3 | 12261 | -0.776 | -0.0406 | No |
| 111 | Myh10 | 12286 | -0.804 | -0.0184 | No |
| 112 | Hadh | 12405 | -1.025 | 0.0029 | No |