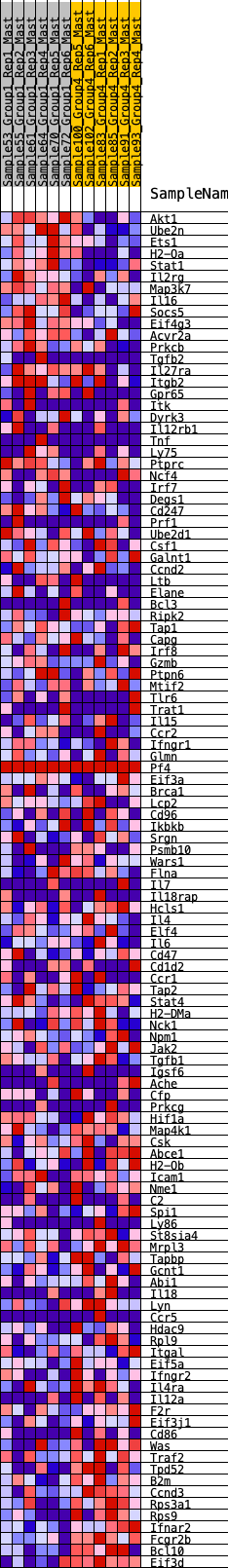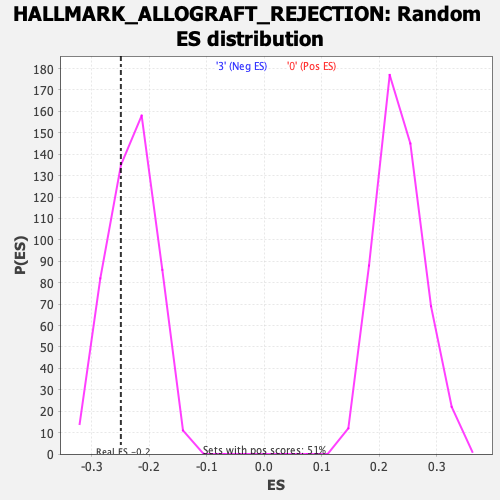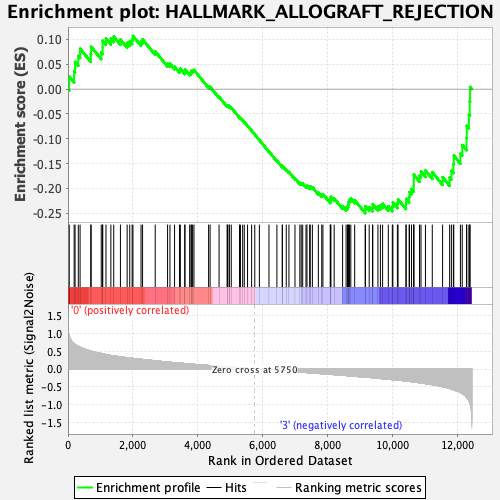
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group4.Mast_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.2497486 |
| Normalized Enrichment Score (NES) | -1.0822184 |
| Nominal p-value | 0.3271605 |
| FDR q-value | 0.5703389 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 38 | 0.920 | 0.0256 | No |
| 2 | Ube2n | 189 | 0.714 | 0.0357 | No |
| 3 | Ets1 | 218 | 0.688 | 0.0548 | No |
| 4 | H2-Oa | 317 | 0.638 | 0.0668 | No |
| 5 | Stat1 | 372 | 0.614 | 0.0815 | No |
| 6 | Il2rg | 702 | 0.507 | 0.0706 | No |
| 7 | Map3k7 | 713 | 0.503 | 0.0855 | No |
| 8 | Il16 | 1024 | 0.438 | 0.0740 | No |
| 9 | Socs5 | 1067 | 0.429 | 0.0839 | No |
| 10 | Eif4g3 | 1068 | 0.429 | 0.0973 | No |
| 11 | Acvr2a | 1170 | 0.407 | 0.1018 | No |
| 12 | Prkcb | 1321 | 0.382 | 0.1015 | No |
| 13 | Tgfb2 | 1409 | 0.370 | 0.1060 | No |
| 14 | Il27ra | 1615 | 0.342 | 0.1000 | No |
| 15 | Itgb2 | 1822 | 0.318 | 0.0932 | No |
| 16 | Gpr65 | 1902 | 0.309 | 0.0964 | No |
| 17 | Itk | 1976 | 0.301 | 0.0998 | No |
| 18 | Dyrk3 | 2001 | 0.299 | 0.1072 | No |
| 19 | Il12rb1 | 2250 | 0.274 | 0.0956 | No |
| 20 | Tnf | 2297 | 0.269 | 0.1003 | No |
| 21 | Ly75 | 2687 | 0.232 | 0.0759 | No |
| 22 | Ptprc | 3068 | 0.195 | 0.0512 | No |
| 23 | Ncf4 | 3141 | 0.188 | 0.0512 | No |
| 24 | Irf7 | 3283 | 0.179 | 0.0453 | No |
| 25 | Degs1 | 3434 | 0.166 | 0.0383 | No |
| 26 | Cd247 | 3461 | 0.164 | 0.0413 | No |
| 27 | Prf1 | 3598 | 0.154 | 0.0351 | No |
| 28 | Ube2d1 | 3603 | 0.153 | 0.0395 | No |
| 29 | Csf1 | 3748 | 0.141 | 0.0323 | No |
| 30 | Galnt1 | 3791 | 0.139 | 0.0332 | No |
| 31 | Ccnd2 | 3800 | 0.138 | 0.0368 | No |
| 32 | Ltb | 3839 | 0.134 | 0.0379 | No |
| 33 | Elane | 3880 | 0.131 | 0.0388 | No |
| 34 | Bcl3 | 4332 | 0.098 | 0.0052 | No |
| 35 | Ripk2 | 4378 | 0.095 | 0.0045 | No |
| 36 | Tap1 | 4654 | 0.075 | -0.0154 | No |
| 37 | Capg | 4902 | 0.057 | -0.0337 | No |
| 38 | Irf8 | 4914 | 0.056 | -0.0329 | No |
| 39 | Gzmb | 4967 | 0.053 | -0.0354 | No |
| 40 | Ptpn6 | 4968 | 0.053 | -0.0338 | No |
| 41 | Mtif2 | 5030 | 0.049 | -0.0372 | No |
| 42 | Tlr6 | 5284 | 0.031 | -0.0568 | No |
| 43 | Trat1 | 5313 | 0.029 | -0.0581 | No |
| 44 | Il15 | 5381 | 0.025 | -0.0628 | No |
| 45 | Ccr2 | 5431 | 0.021 | -0.0661 | No |
| 46 | Ifngr1 | 5532 | 0.015 | -0.0738 | No |
| 47 | Glmn | 5657 | 0.006 | -0.0837 | No |
| 48 | Pf4 | 5754 | 0.000 | -0.0914 | No |
| 49 | Eif3a | 5896 | -0.008 | -0.1026 | No |
| 50 | Brca1 | 6192 | -0.026 | -0.1258 | No |
| 51 | Lcp2 | 6436 | -0.042 | -0.1442 | No |
| 52 | Cd96 | 6602 | -0.052 | -0.1559 | No |
| 53 | Ikbkb | 6604 | -0.052 | -0.1544 | No |
| 54 | Srgn | 6724 | -0.059 | -0.1622 | No |
| 55 | Psmb10 | 6807 | -0.065 | -0.1668 | No |
| 56 | Wars1 | 6993 | -0.077 | -0.1794 | No |
| 57 | Flna | 7143 | -0.086 | -0.1888 | No |
| 58 | Il7 | 7185 | -0.089 | -0.1894 | No |
| 59 | Il18rap | 7231 | -0.093 | -0.1902 | No |
| 60 | Hcls1 | 7332 | -0.099 | -0.1952 | No |
| 61 | Il4 | 7363 | -0.101 | -0.1945 | No |
| 62 | Elf4 | 7446 | -0.107 | -0.1978 | No |
| 63 | Il6 | 7463 | -0.108 | -0.1957 | No |
| 64 | Cd47 | 7529 | -0.113 | -0.1975 | No |
| 65 | Cd1d2 | 7713 | -0.125 | -0.2084 | No |
| 66 | Ccr1 | 7818 | -0.132 | -0.2127 | No |
| 67 | Tap2 | 7853 | -0.134 | -0.2113 | No |
| 68 | Stat4 | 8079 | -0.150 | -0.2249 | No |
| 69 | H2-DMa | 8091 | -0.151 | -0.2211 | No |
| 70 | Nck1 | 8098 | -0.152 | -0.2168 | No |
| 71 | Npm1 | 8200 | -0.158 | -0.2201 | No |
| 72 | Jak2 | 8460 | -0.176 | -0.2356 | No |
| 73 | Tgfb1 | 8568 | -0.184 | -0.2386 | No |
| 74 | Igsf6 | 8612 | -0.187 | -0.2362 | No |
| 75 | Ache | 8637 | -0.189 | -0.2323 | No |
| 76 | Cfp | 8648 | -0.190 | -0.2272 | No |
| 77 | Prkcg | 8671 | -0.191 | -0.2230 | No |
| 78 | Hif1a | 8713 | -0.194 | -0.2203 | No |
| 79 | Map4k1 | 8832 | -0.205 | -0.2235 | No |
| 80 | Csk | 9157 | -0.227 | -0.2427 | Yes |
| 81 | Abce1 | 9162 | -0.228 | -0.2359 | Yes |
| 82 | H2-Ob | 9279 | -0.237 | -0.2379 | Yes |
| 83 | Icam1 | 9381 | -0.246 | -0.2385 | Yes |
| 84 | Nme1 | 9391 | -0.247 | -0.2315 | Yes |
| 85 | C2 | 9549 | -0.261 | -0.2361 | Yes |
| 86 | Spi1 | 9625 | -0.268 | -0.2339 | Yes |
| 87 | Ly86 | 9693 | -0.273 | -0.2308 | Yes |
| 88 | St8sia4 | 9868 | -0.288 | -0.2359 | Yes |
| 89 | Mrpl3 | 9992 | -0.300 | -0.2366 | Yes |
| 90 | Tapbp | 10009 | -0.301 | -0.2285 | Yes |
| 91 | Gcnt1 | 10148 | -0.312 | -0.2300 | Yes |
| 92 | Abi1 | 10168 | -0.313 | -0.2217 | Yes |
| 93 | Il18 | 10413 | -0.337 | -0.2310 | Yes |
| 94 | Lyn | 10418 | -0.338 | -0.2208 | Yes |
| 95 | Ccr5 | 10510 | -0.349 | -0.2173 | Yes |
| 96 | Hdac9 | 10521 | -0.351 | -0.2072 | Yes |
| 97 | Rpl9 | 10583 | -0.357 | -0.2010 | Yes |
| 98 | Itgal | 10651 | -0.367 | -0.1950 | Yes |
| 99 | Eif5a | 10652 | -0.367 | -0.1836 | Yes |
| 100 | Ifngr2 | 10653 | -0.367 | -0.1722 | Yes |
| 101 | Il4ra | 10831 | -0.388 | -0.1744 | Yes |
| 102 | Il12a | 10875 | -0.394 | -0.1657 | Yes |
| 103 | F2r | 11013 | -0.412 | -0.1639 | Yes |
| 104 | Eif3j1 | 11223 | -0.442 | -0.1671 | Yes |
| 105 | Cd86 | 11541 | -0.501 | -0.1772 | Yes |
| 106 | Was | 11758 | -0.546 | -0.1777 | Yes |
| 107 | Traf2 | 11812 | -0.565 | -0.1644 | Yes |
| 108 | Tpd52 | 11873 | -0.583 | -0.1511 | Yes |
| 109 | B2m | 11885 | -0.587 | -0.1337 | Yes |
| 110 | Ccnd3 | 12090 | -0.661 | -0.1297 | Yes |
| 111 | Rps3a1 | 12146 | -0.694 | -0.1125 | Yes |
| 112 | Rps9 | 12279 | -0.796 | -0.0984 | Yes |
| 113 | Ifnar2 | 12285 | -0.801 | -0.0739 | Yes |
| 114 | Fcgr2b | 12351 | -0.879 | -0.0518 | Yes |
| 115 | Bcl10 | 12375 | -0.930 | -0.0247 | Yes |
| 116 | Eif3d | 12388 | -0.961 | 0.0043 | Yes |