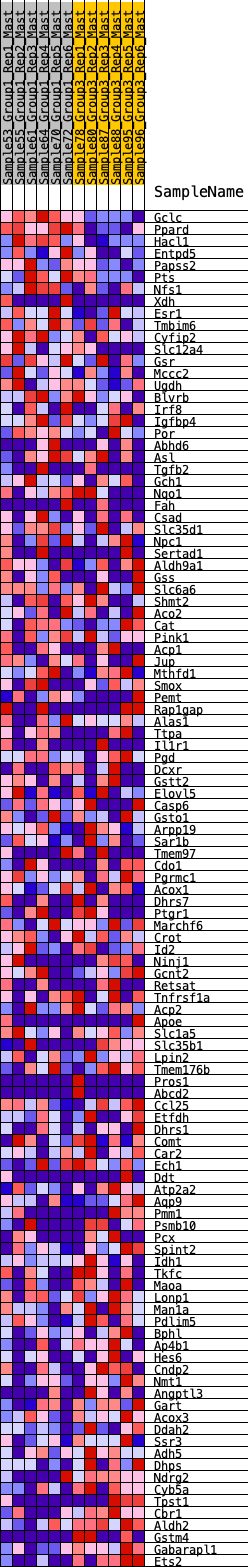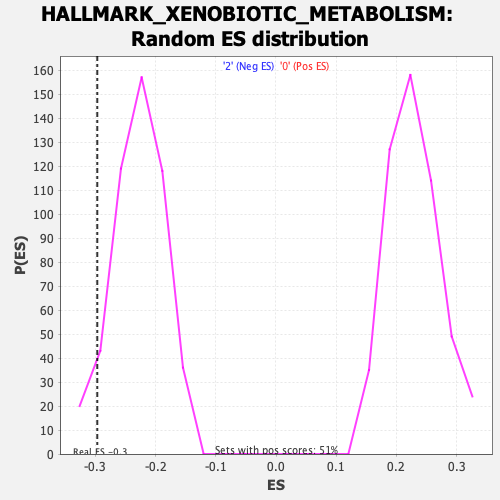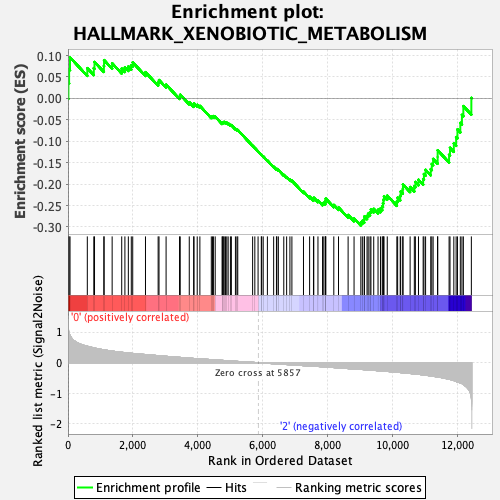
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.29680598 |
| Normalized Enrichment Score (NES) | -1.2994169 |
| Nominal p-value | 0.054766733 |
| FDR q-value | 0.7939469 |
| FWER p-Value | 0.824 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gclc | 13 | 1.123 | 0.0366 | No |
| 2 | Ppard | 34 | 0.976 | 0.0677 | No |
| 3 | Hacl1 | 62 | 0.895 | 0.0956 | No |
| 4 | Entpd5 | 595 | 0.538 | 0.0705 | No |
| 5 | Papss2 | 795 | 0.485 | 0.0706 | No |
| 6 | Pts | 817 | 0.479 | 0.0849 | No |
| 7 | Nfs1 | 1105 | 0.418 | 0.0757 | No |
| 8 | Xdh | 1112 | 0.417 | 0.0892 | No |
| 9 | Esr1 | 1360 | 0.378 | 0.0818 | No |
| 10 | Tmbim6 | 1655 | 0.339 | 0.0693 | No |
| 11 | Cyfip2 | 1753 | 0.327 | 0.0724 | No |
| 12 | Slc12a4 | 1859 | 0.314 | 0.0745 | No |
| 13 | Gsr | 1950 | 0.305 | 0.0774 | No |
| 14 | Mccc2 | 1994 | 0.302 | 0.0840 | No |
| 15 | Ugdh | 2387 | 0.265 | 0.0611 | No |
| 16 | Blvrb | 2777 | 0.229 | 0.0373 | No |
| 17 | Irf8 | 2804 | 0.227 | 0.0427 | No |
| 18 | Igfbp4 | 3022 | 0.208 | 0.0321 | No |
| 19 | Por | 3436 | 0.174 | 0.0044 | No |
| 20 | Abhd6 | 3457 | 0.172 | 0.0086 | No |
| 21 | Asl | 3736 | 0.149 | -0.0090 | No |
| 22 | Tgfb2 | 3871 | 0.138 | -0.0152 | No |
| 23 | Gch1 | 3882 | 0.136 | -0.0115 | No |
| 24 | Nqo1 | 3981 | 0.129 | -0.0151 | No |
| 25 | Fah | 4063 | 0.122 | -0.0176 | No |
| 26 | Csad | 4415 | 0.100 | -0.0427 | No |
| 27 | Slc35d1 | 4451 | 0.097 | -0.0423 | No |
| 28 | Npc1 | 4480 | 0.096 | -0.0413 | No |
| 29 | Sertad1 | 4535 | 0.091 | -0.0427 | No |
| 30 | Aldh9a1 | 4747 | 0.075 | -0.0572 | No |
| 31 | Gss | 4762 | 0.074 | -0.0559 | No |
| 32 | Slc6a6 | 4786 | 0.073 | -0.0553 | No |
| 33 | Shmt2 | 4812 | 0.071 | -0.0550 | No |
| 34 | Aco2 | 4851 | 0.068 | -0.0558 | No |
| 35 | Cat | 4892 | 0.064 | -0.0569 | No |
| 36 | Pink1 | 4934 | 0.061 | -0.0582 | No |
| 37 | Acp1 | 5004 | 0.056 | -0.0619 | No |
| 38 | Jup | 5033 | 0.055 | -0.0623 | No |
| 39 | Mthfd1 | 5158 | 0.046 | -0.0709 | No |
| 40 | Smox | 5204 | 0.043 | -0.0731 | No |
| 41 | Pemt | 5230 | 0.041 | -0.0738 | No |
| 42 | Rap1gap | 5688 | 0.011 | -0.1105 | No |
| 43 | Alas1 | 5756 | 0.006 | -0.1157 | No |
| 44 | Ttpa | 5856 | 0.000 | -0.1237 | No |
| 45 | Il1r1 | 5955 | -0.007 | -0.1314 | No |
| 46 | Pgd | 5967 | -0.008 | -0.1320 | No |
| 47 | Dcxr | 6014 | -0.013 | -0.1353 | No |
| 48 | Gstt2 | 6143 | -0.021 | -0.1450 | No |
| 49 | Elovl5 | 6336 | -0.033 | -0.1595 | No |
| 50 | Casp6 | 6422 | -0.039 | -0.1651 | No |
| 51 | Gsto1 | 6435 | -0.040 | -0.1647 | No |
| 52 | Arpp19 | 6481 | -0.043 | -0.1669 | No |
| 53 | Sar1b | 6648 | -0.055 | -0.1785 | No |
| 54 | Tmem97 | 6736 | -0.060 | -0.1836 | No |
| 55 | Cdo1 | 6837 | -0.067 | -0.1894 | No |
| 56 | Pgrmc1 | 6898 | -0.070 | -0.1919 | No |
| 57 | Acox1 | 7255 | -0.096 | -0.2176 | No |
| 58 | Dhrs7 | 7444 | -0.106 | -0.2293 | No |
| 59 | Ptgr1 | 7563 | -0.114 | -0.2350 | No |
| 60 | Marchf6 | 7572 | -0.115 | -0.2318 | No |
| 61 | Crot | 7701 | -0.124 | -0.2380 | No |
| 62 | Id2 | 7842 | -0.133 | -0.2449 | No |
| 63 | Ninj1 | 7881 | -0.136 | -0.2434 | No |
| 64 | Gcnt2 | 7927 | -0.140 | -0.2424 | No |
| 65 | Retsat | 7936 | -0.141 | -0.2383 | No |
| 66 | Tnfrsf1a | 7942 | -0.141 | -0.2340 | No |
| 67 | Acp2 | 8189 | -0.159 | -0.2486 | No |
| 68 | Apoe | 8334 | -0.170 | -0.2546 | No |
| 69 | Slc1a5 | 8630 | -0.193 | -0.2720 | No |
| 70 | Slc35b1 | 8813 | -0.205 | -0.2799 | No |
| 71 | Lpin2 | 9022 | -0.220 | -0.2894 | Yes |
| 72 | Tmem176b | 9068 | -0.223 | -0.2856 | Yes |
| 73 | Pros1 | 9126 | -0.227 | -0.2826 | Yes |
| 74 | Abcd2 | 9132 | -0.227 | -0.2754 | Yes |
| 75 | Ccl25 | 9213 | -0.235 | -0.2740 | Yes |
| 76 | Etfdh | 9248 | -0.237 | -0.2688 | Yes |
| 77 | Dhrs1 | 9305 | -0.241 | -0.2653 | Yes |
| 78 | Comt | 9333 | -0.244 | -0.2593 | Yes |
| 79 | Car2 | 9419 | -0.251 | -0.2578 | Yes |
| 80 | Ech1 | 9552 | -0.263 | -0.2596 | Yes |
| 81 | Ddt | 9627 | -0.268 | -0.2567 | Yes |
| 82 | Atp2a2 | 9681 | -0.272 | -0.2519 | Yes |
| 83 | Aqp9 | 9704 | -0.274 | -0.2445 | Yes |
| 84 | Pmm1 | 9718 | -0.276 | -0.2363 | Yes |
| 85 | Psmb10 | 9736 | -0.277 | -0.2284 | Yes |
| 86 | Pcx | 9836 | -0.285 | -0.2268 | Yes |
| 87 | Spint2 | 10132 | -0.311 | -0.2403 | Yes |
| 88 | Idh1 | 10156 | -0.314 | -0.2316 | Yes |
| 89 | Tkfc | 10235 | -0.322 | -0.2271 | Yes |
| 90 | Maoa | 10249 | -0.324 | -0.2173 | Yes |
| 91 | Lonp1 | 10315 | -0.329 | -0.2116 | Yes |
| 92 | Man1a | 10320 | -0.329 | -0.2009 | Yes |
| 93 | Pdlim5 | 10540 | -0.353 | -0.2068 | Yes |
| 94 | Bphl | 10670 | -0.368 | -0.2049 | Yes |
| 95 | Ap4b1 | 10702 | -0.371 | -0.1950 | Yes |
| 96 | Hes6 | 10798 | -0.381 | -0.1899 | Yes |
| 97 | Cndp2 | 10942 | -0.400 | -0.1881 | Yes |
| 98 | Nmt1 | 10966 | -0.403 | -0.1764 | Yes |
| 99 | Angptl3 | 11017 | -0.409 | -0.1668 | Yes |
| 100 | Gart | 11176 | -0.433 | -0.1651 | Yes |
| 101 | Acox3 | 11209 | -0.438 | -0.1530 | Yes |
| 102 | Ddah2 | 11249 | -0.444 | -0.1412 | Yes |
| 103 | Ssr3 | 11388 | -0.467 | -0.1368 | Yes |
| 104 | Adh5 | 11389 | -0.467 | -0.1211 | Yes |
| 105 | Dhps | 11741 | -0.543 | -0.1314 | Yes |
| 106 | Ndrg2 | 11769 | -0.547 | -0.1152 | Yes |
| 107 | Cyb5a | 11887 | -0.584 | -0.1051 | Yes |
| 108 | Tpst1 | 11958 | -0.614 | -0.0902 | Yes |
| 109 | Cbr1 | 11999 | -0.630 | -0.0723 | Yes |
| 110 | Aldh2 | 12087 | -0.661 | -0.0572 | Yes |
| 111 | Gstm4 | 12137 | -0.682 | -0.0383 | Yes |
| 112 | Gabarapl1 | 12181 | -0.709 | -0.0180 | Yes |
| 113 | Ets2 | 12427 | -1.164 | 0.0011 | Yes |