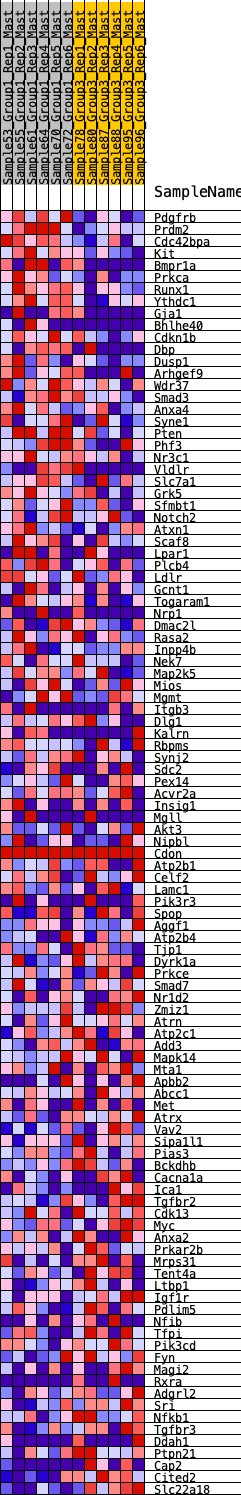
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
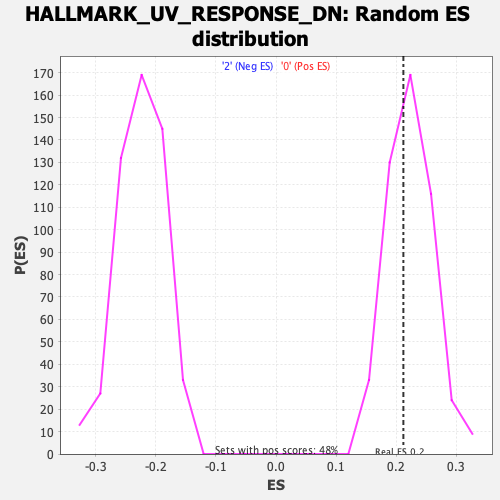
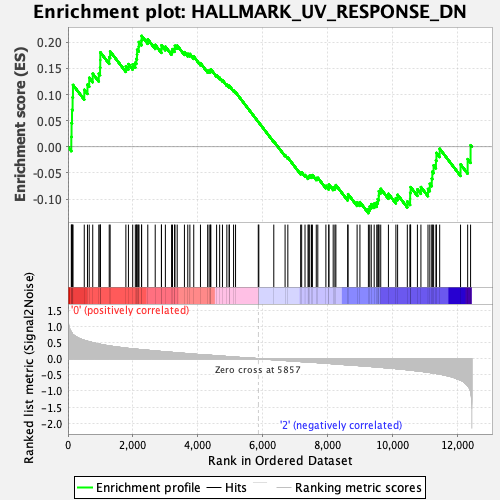
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.21213317 |
| Normalized Enrichment Score (NES) | 0.9463093 |
| Nominal p-value | 0.5987526 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pdgfrb | 102 | 0.821 | 0.0192 | Yes |
| 2 | Prdm2 | 110 | 0.804 | 0.0455 | Yes |
| 3 | Cdc42bpa | 124 | 0.788 | 0.0708 | Yes |
| 4 | Kit | 142 | 0.750 | 0.0945 | Yes |
| 5 | Bmpr1a | 153 | 0.737 | 0.1184 | Yes |
| 6 | Prkca | 501 | 0.563 | 0.1091 | Yes |
| 7 | Runx1 | 600 | 0.536 | 0.1191 | Yes |
| 8 | Ythdc1 | 654 | 0.518 | 0.1321 | Yes |
| 9 | Gja1 | 762 | 0.492 | 0.1399 | Yes |
| 10 | Bhlhe40 | 949 | 0.451 | 0.1399 | Yes |
| 11 | Cdkn1b | 988 | 0.444 | 0.1516 | Yes |
| 12 | Dbp | 994 | 0.442 | 0.1660 | Yes |
| 13 | Dusp1 | 996 | 0.442 | 0.1807 | Yes |
| 14 | Arhgef9 | 1270 | 0.393 | 0.1717 | Yes |
| 15 | Wdr37 | 1298 | 0.387 | 0.1824 | Yes |
| 16 | Smad3 | 1783 | 0.324 | 0.1540 | Yes |
| 17 | Anxa4 | 1862 | 0.314 | 0.1582 | Yes |
| 18 | Syne1 | 1993 | 0.302 | 0.1578 | Yes |
| 19 | Pten | 2074 | 0.294 | 0.1611 | Yes |
| 20 | Phf3 | 2106 | 0.290 | 0.1683 | Yes |
| 21 | Nr3c1 | 2125 | 0.288 | 0.1765 | Yes |
| 22 | Vldlr | 2130 | 0.288 | 0.1858 | Yes |
| 23 | Slc7a1 | 2179 | 0.282 | 0.1913 | Yes |
| 24 | Grk5 | 2183 | 0.282 | 0.2005 | Yes |
| 25 | Sfmbt1 | 2264 | 0.276 | 0.2033 | Yes |
| 26 | Notch2 | 2269 | 0.275 | 0.2121 | Yes |
| 27 | Atxn1 | 2458 | 0.258 | 0.2055 | No |
| 28 | Scaf8 | 2687 | 0.238 | 0.1950 | No |
| 29 | Lpar1 | 2878 | 0.219 | 0.1869 | No |
| 30 | Plcb4 | 2880 | 0.219 | 0.1942 | No |
| 31 | Ldlr | 3000 | 0.210 | 0.1915 | No |
| 32 | Gcnt1 | 3191 | 0.193 | 0.1826 | No |
| 33 | Togaram1 | 3219 | 0.191 | 0.1868 | No |
| 34 | Nrp1 | 3284 | 0.186 | 0.1878 | No |
| 35 | Dmac2l | 3294 | 0.185 | 0.1933 | No |
| 36 | Rasa2 | 3364 | 0.179 | 0.1936 | No |
| 37 | Inpp4b | 3587 | 0.161 | 0.1810 | No |
| 38 | Nek7 | 3690 | 0.153 | 0.1779 | No |
| 39 | Map2k5 | 3755 | 0.148 | 0.1776 | No |
| 40 | Mios | 3874 | 0.137 | 0.1726 | No |
| 41 | Mgmt | 4081 | 0.121 | 0.1600 | No |
| 42 | Itgb3 | 4303 | 0.107 | 0.1457 | No |
| 43 | Dlg1 | 4349 | 0.104 | 0.1455 | No |
| 44 | Kalrn | 4390 | 0.102 | 0.1456 | No |
| 45 | Rbpms | 4401 | 0.101 | 0.1482 | No |
| 46 | Synj2 | 4578 | 0.087 | 0.1368 | No |
| 47 | Sdc2 | 4672 | 0.080 | 0.1320 | No |
| 48 | Pex14 | 4759 | 0.075 | 0.1275 | No |
| 49 | Acvr2a | 4894 | 0.064 | 0.1188 | No |
| 50 | Insig1 | 4967 | 0.058 | 0.1149 | No |
| 51 | Mgll | 4968 | 0.058 | 0.1168 | No |
| 52 | Akt3 | 5101 | 0.050 | 0.1078 | No |
| 53 | Nipbl | 5162 | 0.045 | 0.1044 | No |
| 54 | Cdon | 5862 | 0.000 | 0.0478 | No |
| 55 | Atp2b1 | 5879 | -0.000 | 0.0465 | No |
| 56 | Celf2 | 6340 | -0.034 | 0.0103 | No |
| 57 | Lamc1 | 6688 | -0.057 | -0.0159 | No |
| 58 | Pik3r3 | 6773 | -0.062 | -0.0206 | No |
| 59 | Spop | 7168 | -0.090 | -0.0495 | No |
| 60 | Aggf1 | 7199 | -0.092 | -0.0489 | No |
| 61 | Atp2b4 | 7302 | -0.098 | -0.0539 | No |
| 62 | Tjp1 | 7395 | -0.103 | -0.0579 | No |
| 63 | Dyrk1a | 7417 | -0.104 | -0.0561 | No |
| 64 | Prkce | 7447 | -0.106 | -0.0549 | No |
| 65 | Smad7 | 7502 | -0.110 | -0.0556 | No |
| 66 | Nr1d2 | 7530 | -0.112 | -0.0540 | No |
| 67 | Zmiz1 | 7649 | -0.120 | -0.0596 | No |
| 68 | Atrn | 7696 | -0.124 | -0.0592 | No |
| 69 | Atp2c1 | 7945 | -0.141 | -0.0746 | No |
| 70 | Add3 | 8034 | -0.147 | -0.0768 | No |
| 71 | Mapk14 | 8038 | -0.148 | -0.0721 | No |
| 72 | Mta1 | 8170 | -0.158 | -0.0774 | No |
| 73 | Apbb2 | 8223 | -0.162 | -0.0762 | No |
| 74 | Abcc1 | 8253 | -0.164 | -0.0731 | No |
| 75 | Met | 8617 | -0.191 | -0.0961 | No |
| 76 | Atrx | 8628 | -0.193 | -0.0905 | No |
| 77 | Vav2 | 8907 | -0.211 | -0.1060 | No |
| 78 | Sipa1l1 | 8994 | -0.217 | -0.1057 | No |
| 79 | Pias3 | 9255 | -0.238 | -0.1188 | No |
| 80 | Bckdhb | 9296 | -0.240 | -0.1141 | No |
| 81 | Cacna1a | 9343 | -0.244 | -0.1096 | No |
| 82 | Ica1 | 9433 | -0.251 | -0.1084 | No |
| 83 | Tgfbr2 | 9513 | -0.259 | -0.1062 | No |
| 84 | Cdk13 | 9545 | -0.262 | -0.0999 | No |
| 85 | Myc | 9576 | -0.265 | -0.0935 | No |
| 86 | Anxa2 | 9578 | -0.265 | -0.0847 | No |
| 87 | Prkar2b | 9635 | -0.268 | -0.0803 | No |
| 88 | Mrps31 | 9872 | -0.288 | -0.0898 | No |
| 89 | Tent4a | 10102 | -0.308 | -0.0980 | No |
| 90 | Ltbp1 | 10155 | -0.314 | -0.0917 | No |
| 91 | Igf1r | 10454 | -0.343 | -0.1044 | No |
| 92 | Pdlim5 | 10540 | -0.353 | -0.0995 | No |
| 93 | Nfib | 10542 | -0.353 | -0.0878 | No |
| 94 | Tfpi | 10556 | -0.354 | -0.0770 | No |
| 95 | Pik3cd | 10765 | -0.377 | -0.0812 | No |
| 96 | Fyn | 10873 | -0.390 | -0.0769 | No |
| 97 | Magi2 | 11091 | -0.420 | -0.0804 | No |
| 98 | Rxra | 11148 | -0.429 | -0.0706 | No |
| 99 | Adgrl2 | 11208 | -0.438 | -0.0608 | No |
| 100 | Sri | 11225 | -0.441 | -0.0473 | No |
| 101 | Nfkb1 | 11264 | -0.447 | -0.0355 | No |
| 102 | Tgfbr3 | 11336 | -0.460 | -0.0259 | No |
| 103 | Ddah1 | 11351 | -0.462 | -0.0115 | No |
| 104 | Ptpn21 | 11451 | -0.475 | -0.0037 | No |
| 105 | Cap2 | 12094 | -0.662 | -0.0336 | No |
| 106 | Cited2 | 12313 | -0.830 | -0.0235 | No |
| 107 | Slc22a18 | 12403 | -1.010 | 0.0031 | No |