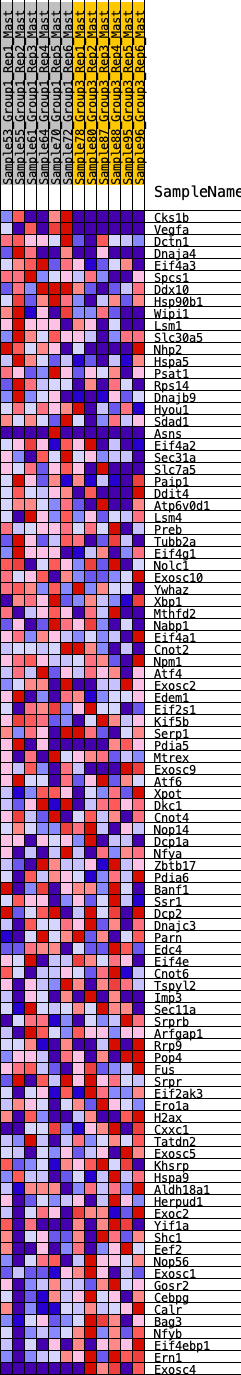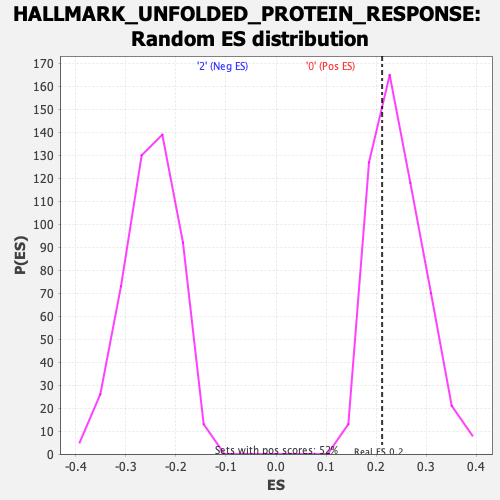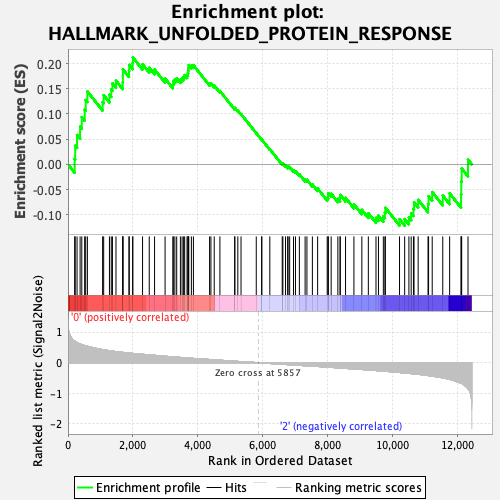
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.2121677 |
| Normalized Enrichment Score (NES) | 0.8706173 |
| Nominal p-value | 0.697318 |
| FDR q-value | 0.9054287 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cks1b | 205 | 0.707 | 0.0111 | Yes |
| 2 | Vegfa | 222 | 0.695 | 0.0371 | Yes |
| 3 | Dctn1 | 279 | 0.658 | 0.0583 | Yes |
| 4 | Dnaja4 | 373 | 0.612 | 0.0748 | Yes |
| 5 | Eif4a3 | 425 | 0.590 | 0.0938 | Yes |
| 6 | Spcs1 | 513 | 0.561 | 0.1087 | Yes |
| 7 | Ddx10 | 545 | 0.550 | 0.1277 | Yes |
| 8 | Hsp90b1 | 597 | 0.537 | 0.1446 | Yes |
| 9 | Wipi1 | 1066 | 0.426 | 0.1234 | Yes |
| 10 | Lsm1 | 1099 | 0.418 | 0.1372 | Yes |
| 11 | Slc30a5 | 1277 | 0.391 | 0.1382 | Yes |
| 12 | Nhp2 | 1333 | 0.382 | 0.1487 | Yes |
| 13 | Hspa5 | 1369 | 0.377 | 0.1606 | Yes |
| 14 | Psat1 | 1478 | 0.362 | 0.1661 | Yes |
| 15 | Rps14 | 1687 | 0.336 | 0.1624 | Yes |
| 16 | Dnajb9 | 1690 | 0.336 | 0.1754 | Yes |
| 17 | Hyou1 | 1691 | 0.336 | 0.1885 | Yes |
| 18 | Sdad1 | 1879 | 0.312 | 0.1856 | Yes |
| 19 | Asns | 1886 | 0.311 | 0.1973 | Yes |
| 20 | Eif4a2 | 1992 | 0.302 | 0.2007 | Yes |
| 21 | Sec31a | 1997 | 0.302 | 0.2122 | Yes |
| 22 | Slc7a5 | 2297 | 0.272 | 0.1986 | No |
| 23 | Paip1 | 2503 | 0.253 | 0.1919 | No |
| 24 | Ddit4 | 2667 | 0.240 | 0.1881 | No |
| 25 | Atp6v0d1 | 2991 | 0.211 | 0.1702 | No |
| 26 | Lsm4 | 3231 | 0.190 | 0.1583 | No |
| 27 | Preb | 3244 | 0.189 | 0.1647 | No |
| 28 | Tubb2a | 3292 | 0.185 | 0.1682 | No |
| 29 | Eif4g1 | 3349 | 0.179 | 0.1707 | No |
| 30 | Nolc1 | 3463 | 0.171 | 0.1682 | No |
| 31 | Exosc10 | 3515 | 0.167 | 0.1706 | No |
| 32 | Ywhaz | 3562 | 0.162 | 0.1733 | No |
| 33 | Xbp1 | 3594 | 0.160 | 0.1770 | No |
| 34 | Mthfd2 | 3668 | 0.155 | 0.1772 | No |
| 35 | Nabp1 | 3695 | 0.152 | 0.1810 | No |
| 36 | Eif4a1 | 3703 | 0.152 | 0.1864 | No |
| 37 | Cnot2 | 3712 | 0.151 | 0.1917 | No |
| 38 | Npm1 | 3721 | 0.151 | 0.1970 | No |
| 39 | Atf4 | 3801 | 0.143 | 0.1962 | No |
| 40 | Exosc2 | 3864 | 0.138 | 0.1966 | No |
| 41 | Edem1 | 4362 | 0.103 | 0.1604 | No |
| 42 | Eif2s1 | 4403 | 0.101 | 0.1611 | No |
| 43 | Kif5b | 4505 | 0.094 | 0.1566 | No |
| 44 | Serp1 | 4680 | 0.080 | 0.1456 | No |
| 45 | Pdia5 | 5135 | 0.048 | 0.1107 | No |
| 46 | Mtrex | 5141 | 0.047 | 0.1121 | No |
| 47 | Exosc9 | 5232 | 0.041 | 0.1064 | No |
| 48 | Atf6 | 5331 | 0.033 | 0.0998 | No |
| 49 | Xpot | 5800 | 0.004 | 0.0620 | No |
| 50 | Dkc1 | 5966 | -0.008 | 0.0490 | No |
| 51 | Cnot4 | 5972 | -0.009 | 0.0489 | No |
| 52 | Nop14 | 6218 | -0.026 | 0.0301 | No |
| 53 | Dcp1a | 6600 | -0.051 | 0.0013 | No |
| 54 | Nfya | 6620 | -0.053 | 0.0018 | No |
| 55 | Zbtb17 | 6702 | -0.058 | -0.0025 | No |
| 56 | Pdia6 | 6769 | -0.062 | -0.0054 | No |
| 57 | Banf1 | 6779 | -0.063 | -0.0037 | No |
| 58 | Ssr1 | 6829 | -0.066 | -0.0051 | No |
| 59 | Dcp2 | 6947 | -0.074 | -0.0117 | No |
| 60 | Dnajc3 | 7008 | -0.078 | -0.0135 | No |
| 61 | Parn | 7128 | -0.087 | -0.0197 | No |
| 62 | Edc4 | 7305 | -0.098 | -0.0301 | No |
| 63 | Eif4e | 7361 | -0.101 | -0.0306 | No |
| 64 | Cnot6 | 7529 | -0.112 | -0.0397 | No |
| 65 | Tspyl2 | 7691 | -0.123 | -0.0479 | No |
| 66 | Imp3 | 7986 | -0.144 | -0.0661 | No |
| 67 | Sec11a | 8016 | -0.146 | -0.0627 | No |
| 68 | Srprb | 8018 | -0.146 | -0.0571 | No |
| 69 | Arfgap1 | 8108 | -0.153 | -0.0583 | No |
| 70 | Rrp9 | 8314 | -0.168 | -0.0683 | No |
| 71 | Pop4 | 8382 | -0.173 | -0.0669 | No |
| 72 | Fus | 8388 | -0.174 | -0.0605 | No |
| 73 | Srpr | 8551 | -0.187 | -0.0663 | No |
| 74 | Eif2ak3 | 8809 | -0.204 | -0.0791 | No |
| 75 | Ero1a | 9053 | -0.222 | -0.0901 | No |
| 76 | H2ax | 9253 | -0.237 | -0.0970 | No |
| 77 | Cxxc1 | 9489 | -0.257 | -0.1060 | No |
| 78 | Tatdn2 | 9565 | -0.264 | -0.1017 | No |
| 79 | Exosc5 | 9715 | -0.275 | -0.1030 | No |
| 80 | Khsrp | 9763 | -0.279 | -0.0958 | No |
| 81 | Hspa9 | 9778 | -0.280 | -0.0860 | No |
| 82 | Aldh18a1 | 10214 | -0.320 | -0.1087 | No |
| 83 | Herpud1 | 10372 | -0.334 | -0.1083 | No |
| 84 | Exoc2 | 10503 | -0.348 | -0.1052 | No |
| 85 | Yif1a | 10573 | -0.357 | -0.0968 | No |
| 86 | Shc1 | 10643 | -0.365 | -0.0881 | No |
| 87 | Eef2 | 10661 | -0.367 | -0.0751 | No |
| 88 | Nop56 | 10788 | -0.380 | -0.0704 | No |
| 89 | Exosc1 | 11092 | -0.421 | -0.0784 | No |
| 90 | Gosr2 | 11109 | -0.423 | -0.0632 | No |
| 91 | Cebpg | 11221 | -0.440 | -0.0549 | No |
| 92 | Calr | 11546 | -0.496 | -0.0617 | No |
| 93 | Bag3 | 11758 | -0.545 | -0.0574 | No |
| 94 | Nfyb | 12110 | -0.670 | -0.0596 | No |
| 95 | Eif4ebp1 | 12116 | -0.672 | -0.0337 | No |
| 96 | Ern1 | 12129 | -0.678 | -0.0081 | No |
| 97 | Exosc4 | 12324 | -0.849 | 0.0095 | No |