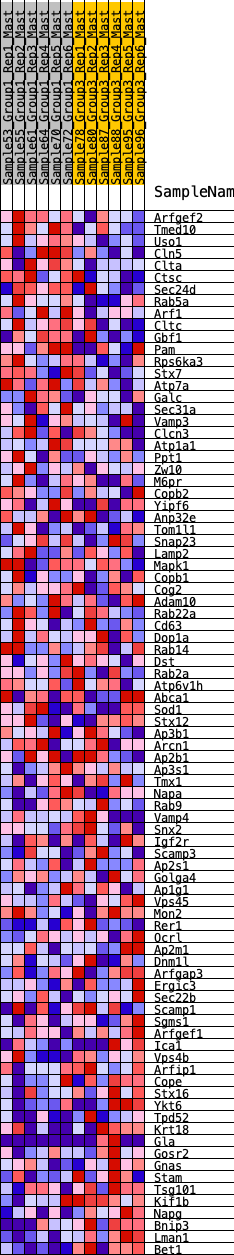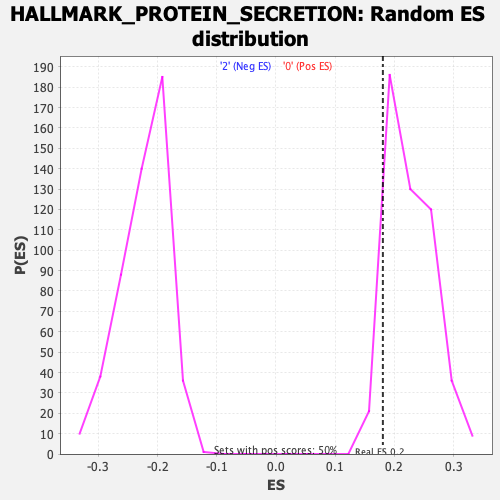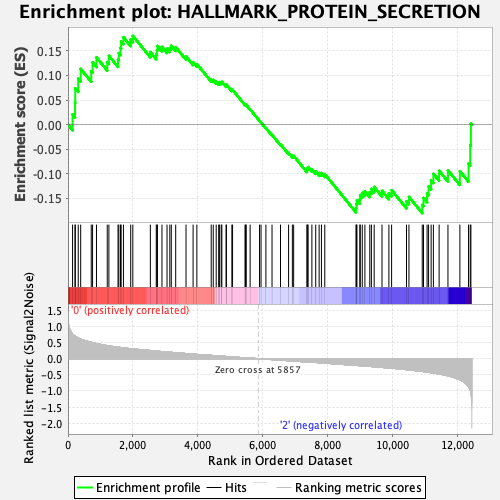
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.18044096 |
| Normalized Enrichment Score (NES) | 0.7957692 |
| Nominal p-value | 0.9063745 |
| FDR q-value | 0.95602757 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Arfgef2 | 141 | 0.751 | 0.0207 | Yes |
| 2 | Tmed10 | 217 | 0.698 | 0.0445 | Yes |
| 3 | Uso1 | 223 | 0.695 | 0.0738 | Yes |
| 4 | Cln5 | 315 | 0.636 | 0.0937 | Yes |
| 5 | Clta | 391 | 0.603 | 0.1134 | Yes |
| 6 | Ctsc | 716 | 0.505 | 0.1087 | Yes |
| 7 | Sec24d | 757 | 0.493 | 0.1266 | Yes |
| 8 | Rab5a | 877 | 0.464 | 0.1368 | Yes |
| 9 | Arf1 | 1209 | 0.402 | 0.1272 | Yes |
| 10 | Cltc | 1260 | 0.394 | 0.1400 | Yes |
| 11 | Gbf1 | 1541 | 0.353 | 0.1324 | Yes |
| 12 | Pam | 1566 | 0.349 | 0.1454 | Yes |
| 13 | Rps6ka3 | 1617 | 0.345 | 0.1561 | Yes |
| 14 | Stx7 | 1637 | 0.341 | 0.1691 | Yes |
| 15 | Atp7a | 1708 | 0.332 | 0.1777 | Yes |
| 16 | Galc | 1932 | 0.306 | 0.1727 | Yes |
| 17 | Sec31a | 1997 | 0.302 | 0.1804 | Yes |
| 18 | Vamp3 | 2537 | 0.251 | 0.1475 | No |
| 19 | Clcn3 | 2719 | 0.235 | 0.1429 | No |
| 20 | Atp1a1 | 2741 | 0.233 | 0.1512 | No |
| 21 | Ppt1 | 2755 | 0.231 | 0.1600 | No |
| 22 | Zw10 | 2895 | 0.218 | 0.1581 | No |
| 23 | M6pr | 3051 | 0.205 | 0.1543 | No |
| 24 | Copb2 | 3137 | 0.198 | 0.1559 | No |
| 25 | Yipf6 | 3182 | 0.194 | 0.1606 | No |
| 26 | Anp32e | 3318 | 0.183 | 0.1575 | No |
| 27 | Tom1l1 | 3636 | 0.157 | 0.1386 | No |
| 28 | Snap23 | 3854 | 0.139 | 0.1270 | No |
| 29 | Lamp2 | 3970 | 0.129 | 0.1232 | No |
| 30 | Mapk1 | 4412 | 0.100 | 0.0918 | No |
| 31 | Copb1 | 4475 | 0.096 | 0.0909 | No |
| 32 | Cog2 | 4566 | 0.088 | 0.0874 | No |
| 33 | Adam10 | 4643 | 0.082 | 0.0847 | No |
| 34 | Rab22a | 4664 | 0.081 | 0.0866 | No |
| 35 | Cd63 | 4719 | 0.077 | 0.0855 | No |
| 36 | Dop1a | 4740 | 0.076 | 0.0871 | No |
| 37 | Rab14 | 4871 | 0.065 | 0.0794 | No |
| 38 | Dst | 4880 | 0.065 | 0.0815 | No |
| 39 | Rab2a | 5049 | 0.053 | 0.0702 | No |
| 40 | Atp6v1h | 5070 | 0.052 | 0.0708 | No |
| 41 | Abca1 | 5454 | 0.025 | 0.0409 | No |
| 42 | Sod1 | 5468 | 0.024 | 0.0408 | No |
| 43 | Stx12 | 5492 | 0.022 | 0.0399 | No |
| 44 | Ap3b1 | 5611 | 0.015 | 0.0310 | No |
| 45 | Arcn1 | 5901 | -0.002 | 0.0077 | No |
| 46 | Ap2b1 | 5947 | -0.006 | 0.0043 | No |
| 47 | Ap3s1 | 6097 | -0.017 | -0.0070 | No |
| 48 | Tmx1 | 6286 | -0.031 | -0.0209 | No |
| 49 | Napa | 6547 | -0.047 | -0.0399 | No |
| 50 | Rab9 | 6796 | -0.064 | -0.0572 | No |
| 51 | Vamp4 | 6912 | -0.071 | -0.0635 | No |
| 52 | Snx2 | 6952 | -0.074 | -0.0635 | No |
| 53 | Igf2r | 7360 | -0.101 | -0.0921 | No |
| 54 | Scamp3 | 7374 | -0.102 | -0.0888 | No |
| 55 | Ap2s1 | 7398 | -0.103 | -0.0863 | No |
| 56 | Golga4 | 7514 | -0.111 | -0.0908 | No |
| 57 | Ap1g1 | 7632 | -0.119 | -0.0952 | No |
| 58 | Vps45 | 7737 | -0.126 | -0.0982 | No |
| 59 | Mon2 | 7808 | -0.131 | -0.0983 | No |
| 60 | Rer1 | 7911 | -0.139 | -0.1006 | No |
| 61 | Ocrl | 8873 | -0.209 | -0.1695 | No |
| 62 | Ap2m1 | 8887 | -0.210 | -0.1616 | No |
| 63 | Dnm1l | 8902 | -0.211 | -0.1537 | No |
| 64 | Arfgap3 | 8992 | -0.216 | -0.1516 | No |
| 65 | Ergic3 | 9003 | -0.218 | -0.1431 | No |
| 66 | Sec22b | 9066 | -0.223 | -0.1386 | No |
| 67 | Scamp1 | 9147 | -0.229 | -0.1353 | No |
| 68 | Sgms1 | 9298 | -0.240 | -0.1372 | No |
| 69 | Arfgef1 | 9347 | -0.245 | -0.1306 | No |
| 70 | Ica1 | 9433 | -0.251 | -0.1267 | No |
| 71 | Vps4b | 9674 | -0.271 | -0.1346 | No |
| 72 | Arfip1 | 9886 | -0.290 | -0.1393 | No |
| 73 | Cope | 9970 | -0.298 | -0.1332 | No |
| 74 | Stx16 | 10429 | -0.340 | -0.1558 | No |
| 75 | Ykt6 | 10504 | -0.348 | -0.1469 | No |
| 76 | Tpd52 | 10914 | -0.396 | -0.1630 | No |
| 77 | Krt18 | 10953 | -0.401 | -0.1490 | No |
| 78 | Gla | 11063 | -0.415 | -0.1400 | No |
| 79 | Gosr2 | 11109 | -0.423 | -0.1256 | No |
| 80 | Gnas | 11188 | -0.434 | -0.1133 | No |
| 81 | Stam | 11257 | -0.446 | -0.0998 | No |
| 82 | Tsg101 | 11434 | -0.473 | -0.0938 | No |
| 83 | Kif1b | 11706 | -0.532 | -0.0930 | No |
| 84 | Napg | 12074 | -0.658 | -0.0946 | No |
| 85 | Bnip3 | 12340 | -0.869 | -0.0789 | No |
| 86 | Lman1 | 12392 | -0.978 | -0.0412 | No |
| 87 | Bet1 | 12413 | -1.054 | 0.0023 | No |