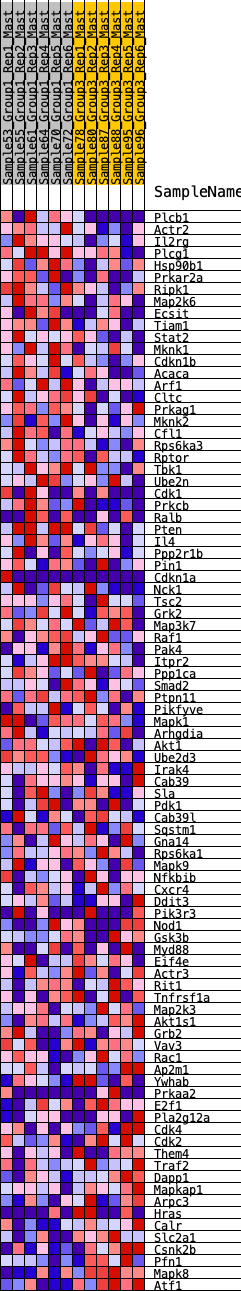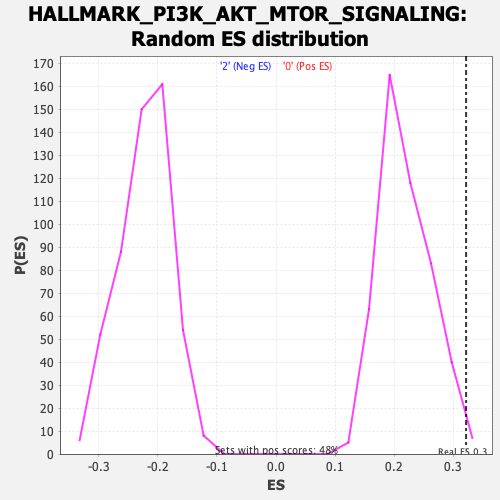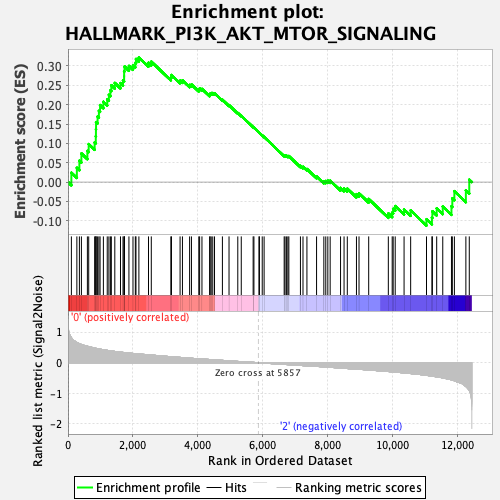
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.32210192 |
| Normalized Enrichment Score (NES) | 1.4669349 |
| Nominal p-value | 0.008316008 |
| FDR q-value | 0.4177672 |
| FWER p-Value | 0.369 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plcb1 | 104 | 0.820 | 0.0242 | Yes |
| 2 | Actr2 | 273 | 0.661 | 0.0369 | Yes |
| 3 | Il2rg | 351 | 0.619 | 0.0553 | Yes |
| 4 | Plcg1 | 413 | 0.595 | 0.0740 | Yes |
| 5 | Hsp90b1 | 597 | 0.537 | 0.0805 | Yes |
| 6 | Prkar2a | 638 | 0.524 | 0.0981 | Yes |
| 7 | Ripk1 | 821 | 0.478 | 0.1024 | Yes |
| 8 | Map2k6 | 856 | 0.470 | 0.1183 | Yes |
| 9 | Ecsit | 861 | 0.468 | 0.1366 | Yes |
| 10 | Tiam1 | 865 | 0.467 | 0.1549 | Yes |
| 11 | Stat2 | 912 | 0.457 | 0.1694 | Yes |
| 12 | Mknk1 | 952 | 0.450 | 0.1841 | Yes |
| 13 | Cdkn1b | 988 | 0.444 | 0.1989 | Yes |
| 14 | Acaca | 1090 | 0.420 | 0.2075 | Yes |
| 15 | Arf1 | 1209 | 0.402 | 0.2139 | Yes |
| 16 | Cltc | 1260 | 0.394 | 0.2255 | Yes |
| 17 | Prkag1 | 1306 | 0.386 | 0.2372 | Yes |
| 18 | Mknk2 | 1335 | 0.381 | 0.2501 | Yes |
| 19 | Cfl1 | 1442 | 0.367 | 0.2561 | Yes |
| 20 | Rps6ka3 | 1617 | 0.345 | 0.2557 | Yes |
| 21 | Rptor | 1695 | 0.335 | 0.2628 | Yes |
| 22 | Tbk1 | 1729 | 0.330 | 0.2733 | Yes |
| 23 | Ube2n | 1730 | 0.330 | 0.2864 | Yes |
| 24 | Cdk1 | 1741 | 0.328 | 0.2986 | Yes |
| 25 | Prkcb | 1877 | 0.313 | 0.3001 | Yes |
| 26 | Ralb | 2002 | 0.301 | 0.3021 | Yes |
| 27 | Pten | 2074 | 0.294 | 0.3080 | Yes |
| 28 | Il4 | 2094 | 0.292 | 0.3181 | Yes |
| 29 | Ppp2r1b | 2184 | 0.282 | 0.3221 | Yes |
| 30 | Pin1 | 2480 | 0.255 | 0.3084 | No |
| 31 | Cdkn1a | 2566 | 0.248 | 0.3114 | No |
| 32 | Nck1 | 3169 | 0.196 | 0.2704 | No |
| 33 | Tsc2 | 3188 | 0.194 | 0.2766 | No |
| 34 | Grk2 | 3453 | 0.172 | 0.2621 | No |
| 35 | Map3k7 | 3526 | 0.165 | 0.2628 | No |
| 36 | Raf1 | 3747 | 0.149 | 0.2509 | No |
| 37 | Pak4 | 3803 | 0.143 | 0.2522 | No |
| 38 | Itpr2 | 4030 | 0.125 | 0.2389 | No |
| 39 | Ppp1ca | 4055 | 0.123 | 0.2418 | No |
| 40 | Smad2 | 4127 | 0.117 | 0.2407 | No |
| 41 | Ptpn11 | 4363 | 0.103 | 0.2258 | No |
| 42 | Pikfyve | 4379 | 0.102 | 0.2286 | No |
| 43 | Mapk1 | 4412 | 0.100 | 0.2300 | No |
| 44 | Arhgdia | 4460 | 0.097 | 0.2301 | No |
| 45 | Akt1 | 4514 | 0.092 | 0.2294 | No |
| 46 | Ube2d3 | 4757 | 0.075 | 0.2128 | No |
| 47 | Irak4 | 4961 | 0.059 | 0.1987 | No |
| 48 | Cab39 | 5231 | 0.041 | 0.1786 | No |
| 49 | Sla | 5338 | 0.033 | 0.1713 | No |
| 50 | Pdk1 | 5704 | 0.010 | 0.1421 | No |
| 51 | Cab39l | 5728 | 0.009 | 0.1406 | No |
| 52 | Sqstm1 | 5883 | -0.000 | 0.1282 | No |
| 53 | Gna14 | 5898 | -0.002 | 0.1271 | No |
| 54 | Rps6ka1 | 5984 | -0.010 | 0.1206 | No |
| 55 | Mapk9 | 6042 | -0.015 | 0.1166 | No |
| 56 | Nfkbib | 6660 | -0.056 | 0.0688 | No |
| 57 | Cxcr4 | 6686 | -0.057 | 0.0691 | No |
| 58 | Ddit3 | 6729 | -0.060 | 0.0681 | No |
| 59 | Pik3r3 | 6773 | -0.062 | 0.0671 | No |
| 60 | Nod1 | 6804 | -0.065 | 0.0672 | No |
| 61 | Gsk3b | 7161 | -0.090 | 0.0420 | No |
| 62 | Myd88 | 7240 | -0.095 | 0.0394 | No |
| 63 | Eif4e | 7361 | -0.101 | 0.0337 | No |
| 64 | Actr3 | 7658 | -0.120 | 0.0146 | No |
| 65 | Rit1 | 7882 | -0.136 | 0.0019 | No |
| 66 | Tnfrsf1a | 7942 | -0.141 | 0.0028 | No |
| 67 | Map2k3 | 8005 | -0.145 | 0.0035 | No |
| 68 | Akt1s1 | 8077 | -0.150 | 0.0038 | No |
| 69 | Grb2 | 8397 | -0.174 | -0.0151 | No |
| 70 | Vav3 | 8505 | -0.183 | -0.0165 | No |
| 71 | Rac1 | 8605 | -0.191 | -0.0170 | No |
| 72 | Ap2m1 | 8887 | -0.210 | -0.0314 | No |
| 73 | Ywhab | 8966 | -0.215 | -0.0291 | No |
| 74 | Prkaa2 | 9264 | -0.238 | -0.0437 | No |
| 75 | E2f1 | 9869 | -0.288 | -0.0811 | No |
| 76 | Pla2g12a | 9988 | -0.299 | -0.0788 | No |
| 77 | Cdk4 | 10018 | -0.301 | -0.0692 | No |
| 78 | Cdk2 | 10081 | -0.306 | -0.0620 | No |
| 79 | Them4 | 10354 | -0.333 | -0.0708 | No |
| 80 | Traf2 | 10557 | -0.354 | -0.0731 | No |
| 81 | Dapp1 | 11045 | -0.413 | -0.0961 | No |
| 82 | Mapkap1 | 11212 | -0.439 | -0.0921 | No |
| 83 | Arpc3 | 11226 | -0.441 | -0.0756 | No |
| 84 | Hras | 11359 | -0.463 | -0.0679 | No |
| 85 | Calr | 11546 | -0.496 | -0.0632 | No |
| 86 | Slc2a1 | 11814 | -0.561 | -0.0625 | No |
| 87 | Csnk2b | 11841 | -0.569 | -0.0420 | No |
| 88 | Pfn1 | 11904 | -0.590 | -0.0236 | No |
| 89 | Mapk8 | 12258 | -0.777 | -0.0212 | No |
| 90 | Atf1 | 12363 | -0.905 | 0.0063 | No |