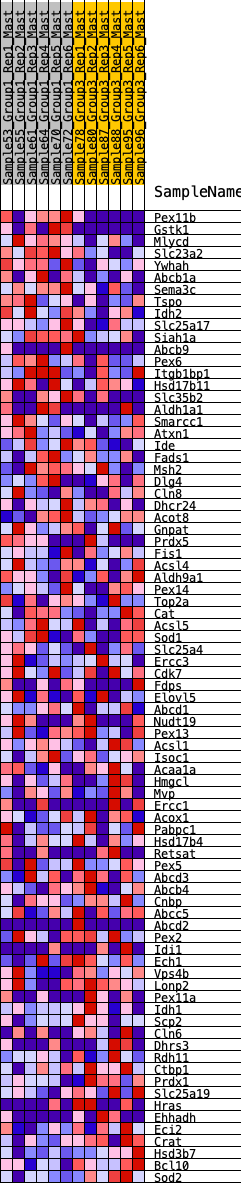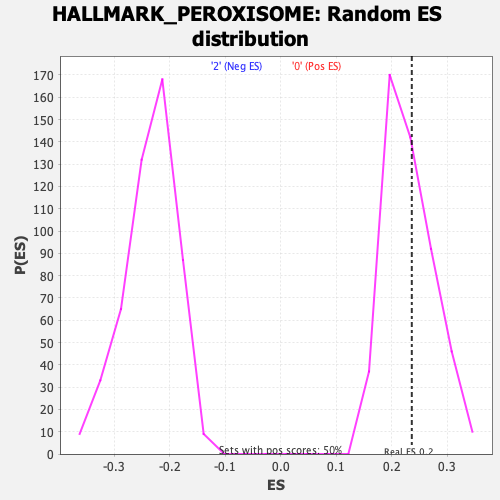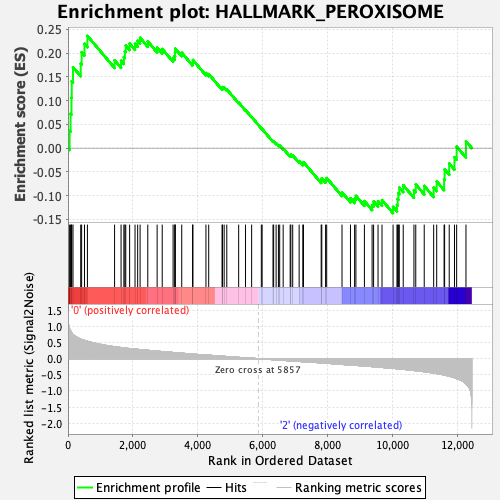
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.23633024 |
| Normalized Enrichment Score (NES) | 1.0217745 |
| Nominal p-value | 0.4305835 |
| FDR q-value | 0.8800564 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex11b | 50 | 0.922 | 0.0367 | Yes |
| 2 | Gstk1 | 79 | 0.859 | 0.0725 | Yes |
| 3 | Mlycd | 107 | 0.810 | 0.1062 | Yes |
| 4 | Slc23a2 | 115 | 0.796 | 0.1409 | Yes |
| 5 | Ywhah | 154 | 0.737 | 0.1704 | Yes |
| 6 | Abcb1a | 392 | 0.603 | 0.1779 | Yes |
| 7 | Sema3c | 420 | 0.593 | 0.2020 | Yes |
| 8 | Tspo | 508 | 0.562 | 0.2198 | Yes |
| 9 | Idh2 | 598 | 0.537 | 0.2363 | Yes |
| 10 | Slc25a17 | 1435 | 0.368 | 0.1850 | No |
| 11 | Siah1a | 1635 | 0.341 | 0.1839 | No |
| 12 | Abcb9 | 1721 | 0.330 | 0.1917 | No |
| 13 | Pex6 | 1752 | 0.327 | 0.2038 | No |
| 14 | Itgb1bp1 | 1779 | 0.324 | 0.2160 | No |
| 15 | Hsd17b11 | 1899 | 0.310 | 0.2201 | No |
| 16 | Slc35b2 | 2065 | 0.295 | 0.2198 | No |
| 17 | Aldh1a1 | 2146 | 0.286 | 0.2260 | No |
| 18 | Smarcc1 | 2221 | 0.280 | 0.2324 | No |
| 19 | Atxn1 | 2458 | 0.258 | 0.2247 | No |
| 20 | Ide | 2746 | 0.232 | 0.2118 | No |
| 21 | Fads1 | 2903 | 0.217 | 0.2088 | No |
| 22 | Msh2 | 3238 | 0.190 | 0.1901 | No |
| 23 | Dlg4 | 3289 | 0.186 | 0.1943 | No |
| 24 | Cln8 | 3299 | 0.185 | 0.2017 | No |
| 25 | Dhcr24 | 3304 | 0.184 | 0.2096 | No |
| 26 | Acot8 | 3504 | 0.168 | 0.2009 | No |
| 27 | Gnpat | 3837 | 0.141 | 0.1803 | No |
| 28 | Prdx5 | 3849 | 0.140 | 0.1855 | No |
| 29 | Fis1 | 4250 | 0.111 | 0.1581 | No |
| 30 | Acsl4 | 4335 | 0.105 | 0.1560 | No |
| 31 | Aldh9a1 | 4747 | 0.075 | 0.1261 | No |
| 32 | Pex14 | 4759 | 0.075 | 0.1285 | No |
| 33 | Top2a | 4811 | 0.071 | 0.1275 | No |
| 34 | Cat | 4892 | 0.064 | 0.1238 | No |
| 35 | Acsl5 | 5258 | 0.039 | 0.0960 | No |
| 36 | Sod1 | 5468 | 0.024 | 0.0802 | No |
| 37 | Slc25a4 | 5658 | 0.013 | 0.0654 | No |
| 38 | Ercc3 | 5956 | -0.007 | 0.0417 | No |
| 39 | Cdk7 | 5981 | -0.009 | 0.0402 | No |
| 40 | Fdps | 6316 | -0.032 | 0.0146 | No |
| 41 | Elovl5 | 6336 | -0.033 | 0.0145 | No |
| 42 | Abcd1 | 6412 | -0.039 | 0.0102 | No |
| 43 | Nudt19 | 6483 | -0.043 | 0.0064 | No |
| 44 | Pex13 | 6522 | -0.046 | 0.0054 | No |
| 45 | Acsl1 | 6629 | -0.053 | -0.0009 | No |
| 46 | Isoc1 | 6845 | -0.067 | -0.0153 | No |
| 47 | Acaa1a | 6866 | -0.069 | -0.0139 | No |
| 48 | Hmgcl | 6923 | -0.072 | -0.0152 | No |
| 49 | Mvp | 7121 | -0.087 | -0.0273 | No |
| 50 | Ercc1 | 7234 | -0.094 | -0.0322 | No |
| 51 | Acox1 | 7255 | -0.096 | -0.0296 | No |
| 52 | Pabpc1 | 7800 | -0.130 | -0.0678 | No |
| 53 | Hsd17b4 | 7819 | -0.132 | -0.0634 | No |
| 54 | Retsat | 7936 | -0.141 | -0.0666 | No |
| 55 | Pex5 | 7970 | -0.143 | -0.0629 | No |
| 56 | Abcd3 | 8442 | -0.179 | -0.0931 | No |
| 57 | Abcb4 | 8703 | -0.197 | -0.1054 | No |
| 58 | Cnbp | 8830 | -0.206 | -0.1065 | No |
| 59 | Abcc5 | 8870 | -0.209 | -0.1004 | No |
| 60 | Abcd2 | 9132 | -0.227 | -0.1115 | No |
| 61 | Pex2 | 9369 | -0.246 | -0.1197 | No |
| 62 | Idi1 | 9415 | -0.250 | -0.1123 | No |
| 63 | Ech1 | 9552 | -0.263 | -0.1116 | No |
| 64 | Vps4b | 9674 | -0.271 | -0.1094 | No |
| 65 | Lonp2 | 10015 | -0.301 | -0.1236 | No |
| 66 | Pex11a | 10135 | -0.312 | -0.1194 | No |
| 67 | Idh1 | 10156 | -0.314 | -0.1071 | No |
| 68 | Scp2 | 10179 | -0.316 | -0.0949 | No |
| 69 | Cln6 | 10207 | -0.319 | -0.0830 | No |
| 70 | Dhrs3 | 10328 | -0.330 | -0.0781 | No |
| 71 | Rdh11 | 10658 | -0.367 | -0.0885 | No |
| 72 | Ctbp1 | 10714 | -0.372 | -0.0764 | No |
| 73 | Prdx1 | 10975 | -0.404 | -0.0796 | No |
| 74 | Slc25a19 | 11266 | -0.447 | -0.0832 | No |
| 75 | Hras | 11359 | -0.463 | -0.0702 | No |
| 76 | Ehhadh | 11587 | -0.505 | -0.0662 | No |
| 77 | Eci2 | 11603 | -0.509 | -0.0449 | No |
| 78 | Crat | 11745 | -0.544 | -0.0322 | No |
| 79 | Hsd3b7 | 11907 | -0.593 | -0.0190 | No |
| 80 | Bcl10 | 11974 | -0.621 | 0.0031 | No |
| 81 | Sod2 | 12261 | -0.781 | 0.0146 | No |