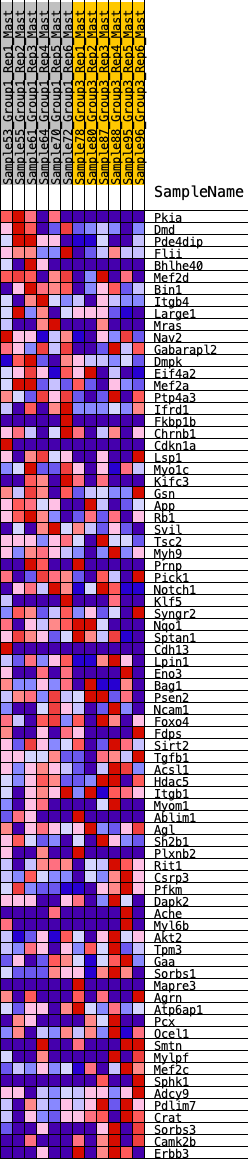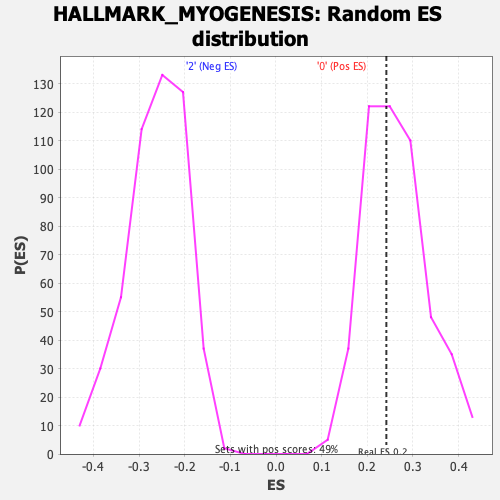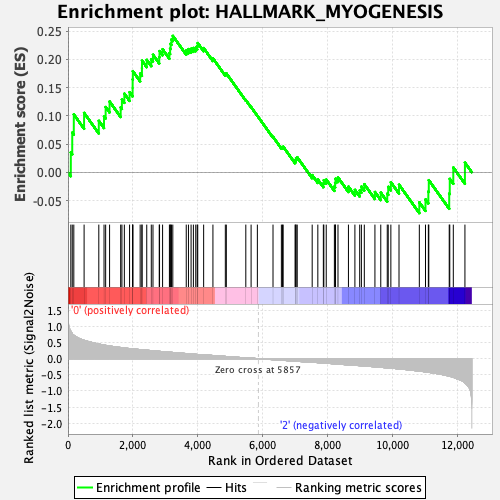
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.24183084 |
| Normalized Enrichment Score (NES) | 0.92155 |
| Nominal p-value | 0.5731707 |
| FDR q-value | 0.9673271 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pkia | 87 | 0.848 | 0.0353 | Yes |
| 2 | Dmd | 131 | 0.774 | 0.0704 | Yes |
| 3 | Pde4dip | 179 | 0.721 | 0.1026 | Yes |
| 4 | Flii | 496 | 0.565 | 0.1053 | Yes |
| 5 | Bhlhe40 | 949 | 0.451 | 0.0913 | Yes |
| 6 | Mef2d | 1110 | 0.417 | 0.0992 | Yes |
| 7 | Bin1 | 1160 | 0.411 | 0.1157 | Yes |
| 8 | Itgb4 | 1281 | 0.390 | 0.1255 | Yes |
| 9 | Large1 | 1623 | 0.344 | 0.1151 | Yes |
| 10 | Mras | 1661 | 0.339 | 0.1290 | Yes |
| 11 | Nav2 | 1736 | 0.329 | 0.1394 | Yes |
| 12 | Gabarapl2 | 1897 | 0.311 | 0.1420 | Yes |
| 13 | Dmpk | 1991 | 0.302 | 0.1495 | Yes |
| 14 | Eif4a2 | 1992 | 0.302 | 0.1646 | Yes |
| 15 | Mef2a | 1998 | 0.301 | 0.1793 | Yes |
| 16 | Ptp4a3 | 2223 | 0.280 | 0.1751 | Yes |
| 17 | Ifrd1 | 2278 | 0.274 | 0.1844 | Yes |
| 18 | Fkbp1b | 2280 | 0.274 | 0.1980 | Yes |
| 19 | Chrnb1 | 2426 | 0.261 | 0.1993 | Yes |
| 20 | Cdkn1a | 2566 | 0.248 | 0.2005 | Yes |
| 21 | Lsp1 | 2618 | 0.243 | 0.2085 | Yes |
| 22 | Myo1c | 2811 | 0.226 | 0.2043 | Yes |
| 23 | Kifc3 | 2820 | 0.225 | 0.2149 | Yes |
| 24 | Gsn | 2914 | 0.216 | 0.2181 | Yes |
| 25 | App | 3124 | 0.199 | 0.2112 | Yes |
| 26 | Rb1 | 3146 | 0.197 | 0.2193 | Yes |
| 27 | Svil | 3162 | 0.196 | 0.2279 | Yes |
| 28 | Tsc2 | 3188 | 0.194 | 0.2356 | Yes |
| 29 | Myh9 | 3229 | 0.190 | 0.2418 | Yes |
| 30 | Prnp | 3643 | 0.157 | 0.2163 | No |
| 31 | Pick1 | 3711 | 0.151 | 0.2184 | No |
| 32 | Notch1 | 3792 | 0.144 | 0.2191 | No |
| 33 | Klf5 | 3863 | 0.139 | 0.2204 | No |
| 34 | Syngr2 | 3936 | 0.132 | 0.2212 | No |
| 35 | Nqo1 | 3981 | 0.129 | 0.2240 | No |
| 36 | Sptan1 | 3997 | 0.127 | 0.2292 | No |
| 37 | Cdh13 | 4179 | 0.114 | 0.2202 | No |
| 38 | Lpin1 | 4465 | 0.096 | 0.2020 | No |
| 39 | Eno3 | 4847 | 0.068 | 0.1746 | No |
| 40 | Bag1 | 4878 | 0.065 | 0.1754 | No |
| 41 | Psen2 | 5478 | 0.023 | 0.1281 | No |
| 42 | Ncam1 | 5641 | 0.013 | 0.1156 | No |
| 43 | Foxo4 | 5834 | 0.002 | 0.1002 | No |
| 44 | Fdps | 6316 | -0.032 | 0.0629 | No |
| 45 | Sirt2 | 6574 | -0.050 | 0.0446 | No |
| 46 | Tgfb1 | 6608 | -0.052 | 0.0445 | No |
| 47 | Acsl1 | 6629 | -0.053 | 0.0455 | No |
| 48 | Hdac5 | 6996 | -0.077 | 0.0198 | No |
| 49 | Itgb1 | 7014 | -0.078 | 0.0223 | No |
| 50 | Myom1 | 7028 | -0.079 | 0.0252 | No |
| 51 | Ablim1 | 7062 | -0.082 | 0.0267 | No |
| 52 | Agl | 7523 | -0.112 | -0.0050 | No |
| 53 | Sh2b1 | 7697 | -0.124 | -0.0128 | No |
| 54 | Plxnb2 | 7867 | -0.136 | -0.0197 | No |
| 55 | Rit1 | 7882 | -0.136 | -0.0140 | No |
| 56 | Csrp3 | 7956 | -0.142 | -0.0128 | No |
| 57 | Pfkm | 8207 | -0.161 | -0.0250 | No |
| 58 | Dapk2 | 8231 | -0.163 | -0.0188 | No |
| 59 | Ache | 8242 | -0.163 | -0.0114 | No |
| 60 | Myl6b | 8318 | -0.168 | -0.0091 | No |
| 61 | Akt2 | 8640 | -0.193 | -0.0254 | No |
| 62 | Tpm3 | 8839 | -0.206 | -0.0311 | No |
| 63 | Gaa | 8987 | -0.216 | -0.0322 | No |
| 64 | Sorbs1 | 9039 | -0.221 | -0.0253 | No |
| 65 | Mapre3 | 9129 | -0.227 | -0.0211 | No |
| 66 | Agrn | 9456 | -0.253 | -0.0349 | No |
| 67 | Atp6ap1 | 9636 | -0.268 | -0.0360 | No |
| 68 | Pcx | 9836 | -0.285 | -0.0378 | No |
| 69 | Ocel1 | 9866 | -0.288 | -0.0258 | No |
| 70 | Smtn | 9947 | -0.295 | -0.0175 | No |
| 71 | Mylpf | 10198 | -0.318 | -0.0219 | No |
| 72 | Mef2c | 10824 | -0.385 | -0.0532 | No |
| 73 | Sphk1 | 11014 | -0.408 | -0.0481 | No |
| 74 | Adcy9 | 11102 | -0.422 | -0.0341 | No |
| 75 | Pdlim7 | 11116 | -0.424 | -0.0140 | No |
| 76 | Crat | 11745 | -0.544 | -0.0376 | No |
| 77 | Sorbs3 | 11762 | -0.546 | -0.0116 | No |
| 78 | Camk2b | 11871 | -0.578 | 0.0085 | No |
| 79 | Erbb3 | 12229 | -0.752 | 0.0171 | No |