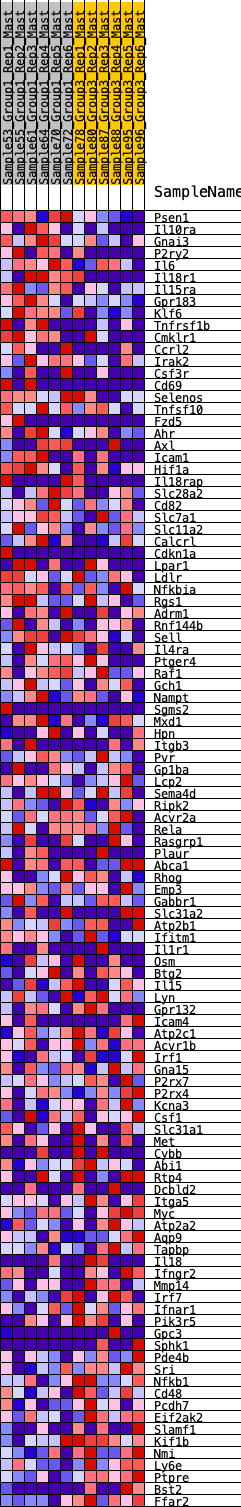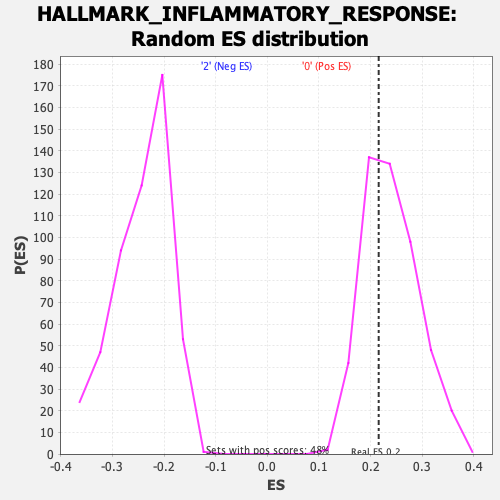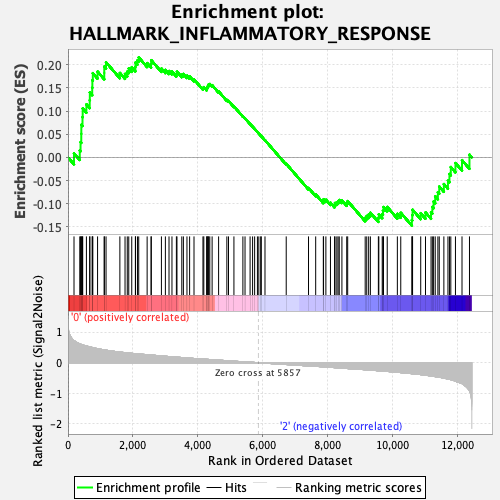
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.21577056 |
| Normalized Enrichment Score (NES) | 0.9012078 |
| Nominal p-value | 0.6307054 |
| FDR q-value | 0.8828878 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Psen1 | 185 | 0.719 | 0.0088 | Yes |
| 2 | Il10ra | 363 | 0.615 | 0.0148 | Yes |
| 3 | Gnai3 | 387 | 0.605 | 0.0329 | Yes |
| 4 | P2ry2 | 409 | 0.596 | 0.0509 | Yes |
| 5 | Il6 | 411 | 0.596 | 0.0705 | Yes |
| 6 | Il18r1 | 447 | 0.581 | 0.0869 | Yes |
| 7 | Il15ra | 453 | 0.579 | 0.1056 | Yes |
| 8 | Gpr183 | 567 | 0.545 | 0.1145 | Yes |
| 9 | Klf6 | 669 | 0.514 | 0.1233 | Yes |
| 10 | Tnfrsf1b | 673 | 0.513 | 0.1400 | Yes |
| 11 | Cmklr1 | 746 | 0.497 | 0.1506 | Yes |
| 12 | Ccrl2 | 751 | 0.494 | 0.1666 | Yes |
| 13 | Irak2 | 765 | 0.492 | 0.1818 | Yes |
| 14 | Csf3r | 911 | 0.457 | 0.1851 | Yes |
| 15 | Cd69 | 1117 | 0.417 | 0.1823 | Yes |
| 16 | Selenos | 1118 | 0.417 | 0.1961 | Yes |
| 17 | Tnfsf10 | 1172 | 0.408 | 0.2052 | Yes |
| 18 | Fzd5 | 1598 | 0.346 | 0.1822 | Yes |
| 19 | Ahr | 1757 | 0.327 | 0.1802 | Yes |
| 20 | Axl | 1826 | 0.318 | 0.1852 | Yes |
| 21 | Icam1 | 1872 | 0.313 | 0.1919 | Yes |
| 22 | Hif1a | 1964 | 0.305 | 0.1946 | Yes |
| 23 | Il18rap | 2076 | 0.294 | 0.1954 | Yes |
| 24 | Slc28a2 | 2080 | 0.294 | 0.2048 | Yes |
| 25 | Cd82 | 2140 | 0.286 | 0.2095 | Yes |
| 26 | Slc7a1 | 2179 | 0.282 | 0.2158 | Yes |
| 27 | Slc11a2 | 2438 | 0.260 | 0.2034 | No |
| 28 | Calcrl | 2559 | 0.249 | 0.2019 | No |
| 29 | Cdkn1a | 2566 | 0.248 | 0.2097 | No |
| 30 | Lpar1 | 2878 | 0.219 | 0.1917 | No |
| 31 | Ldlr | 3000 | 0.210 | 0.1888 | No |
| 32 | Nfkbia | 3112 | 0.200 | 0.1865 | No |
| 33 | Rgs1 | 3203 | 0.192 | 0.1855 | No |
| 34 | Adrm1 | 3339 | 0.181 | 0.1805 | No |
| 35 | Rnf144b | 3357 | 0.179 | 0.1851 | No |
| 36 | Sell | 3506 | 0.168 | 0.1786 | No |
| 37 | Il4ra | 3555 | 0.163 | 0.1801 | No |
| 38 | Ptger4 | 3665 | 0.155 | 0.1764 | No |
| 39 | Raf1 | 3747 | 0.149 | 0.1747 | No |
| 40 | Gch1 | 3882 | 0.136 | 0.1684 | No |
| 41 | Nampt | 4158 | 0.115 | 0.1499 | No |
| 42 | Sgms2 | 4184 | 0.114 | 0.1516 | No |
| 43 | Mxd1 | 4273 | 0.109 | 0.1481 | No |
| 44 | Hpn | 4281 | 0.109 | 0.1511 | No |
| 45 | Itgb3 | 4303 | 0.107 | 0.1530 | No |
| 46 | Pvr | 4317 | 0.107 | 0.1554 | No |
| 47 | Gp1ba | 4334 | 0.105 | 0.1576 | No |
| 48 | Lcp2 | 4368 | 0.103 | 0.1584 | No |
| 49 | Sema4d | 4440 | 0.098 | 0.1558 | No |
| 50 | Ripk2 | 4640 | 0.083 | 0.1424 | No |
| 51 | Acvr2a | 4894 | 0.064 | 0.1240 | No |
| 52 | Rela | 4945 | 0.060 | 0.1219 | No |
| 53 | Rasgrp1 | 5112 | 0.049 | 0.1101 | No |
| 54 | Plaur | 5388 | 0.029 | 0.0888 | No |
| 55 | Abca1 | 5454 | 0.025 | 0.0843 | No |
| 56 | Rhog | 5610 | 0.015 | 0.0723 | No |
| 57 | Emp3 | 5687 | 0.011 | 0.0665 | No |
| 58 | Gabbr1 | 5750 | 0.007 | 0.0617 | No |
| 59 | Slc31a2 | 5836 | 0.002 | 0.0548 | No |
| 60 | Atp2b1 | 5879 | -0.000 | 0.0514 | No |
| 61 | Ifitm1 | 5937 | -0.005 | 0.0470 | No |
| 62 | Il1r1 | 5955 | -0.007 | 0.0459 | No |
| 63 | Osm | 6069 | -0.016 | 0.0372 | No |
| 64 | Btg2 | 6723 | -0.059 | -0.0137 | No |
| 65 | Il15 | 7410 | -0.104 | -0.0659 | No |
| 66 | Lyn | 7631 | -0.119 | -0.0798 | No |
| 67 | Gpr132 | 7866 | -0.136 | -0.0943 | No |
| 68 | Icam4 | 7873 | -0.136 | -0.0903 | No |
| 69 | Atp2c1 | 7945 | -0.141 | -0.0914 | No |
| 70 | Acvr1b | 8086 | -0.152 | -0.0977 | No |
| 71 | Irf1 | 8213 | -0.161 | -0.1026 | No |
| 72 | Gna15 | 8226 | -0.162 | -0.0983 | No |
| 73 | P2rx7 | 8283 | -0.166 | -0.0973 | No |
| 74 | P2rx4 | 8321 | -0.168 | -0.0947 | No |
| 75 | Kcna3 | 8357 | -0.171 | -0.0919 | No |
| 76 | Csf1 | 8439 | -0.178 | -0.0926 | No |
| 77 | Slc31a1 | 8587 | -0.190 | -0.0982 | No |
| 78 | Met | 8617 | -0.191 | -0.0943 | No |
| 79 | Cybb | 9156 | -0.230 | -0.1303 | No |
| 80 | Abi1 | 9204 | -0.234 | -0.1264 | No |
| 81 | Rtp4 | 9263 | -0.238 | -0.1232 | No |
| 82 | Dcbld2 | 9316 | -0.243 | -0.1194 | No |
| 83 | Itga5 | 9562 | -0.264 | -0.1305 | No |
| 84 | Myc | 9576 | -0.265 | -0.1228 | No |
| 85 | Atp2a2 | 9681 | -0.272 | -0.1223 | No |
| 86 | Aqp9 | 9704 | -0.274 | -0.1150 | No |
| 87 | Tapbp | 9720 | -0.276 | -0.1071 | No |
| 88 | Il18 | 9835 | -0.285 | -0.1069 | No |
| 89 | Ifngr2 | 10147 | -0.313 | -0.1218 | No |
| 90 | Mmp14 | 10252 | -0.324 | -0.1195 | No |
| 91 | Irf7 | 10594 | -0.359 | -0.1353 | No |
| 92 | Ifnar1 | 10607 | -0.361 | -0.1243 | No |
| 93 | Pik3r5 | 10616 | -0.362 | -0.1130 | No |
| 94 | Gpc3 | 10867 | -0.389 | -0.1204 | No |
| 95 | Sphk1 | 11014 | -0.408 | -0.1188 | No |
| 96 | Pde4b | 11185 | -0.434 | -0.1182 | No |
| 97 | Sri | 11225 | -0.441 | -0.1068 | No |
| 98 | Nfkb1 | 11264 | -0.447 | -0.0951 | No |
| 99 | Cd48 | 11314 | -0.455 | -0.0840 | No |
| 100 | Pcdh7 | 11396 | -0.467 | -0.0751 | No |
| 101 | Eif2ak2 | 11438 | -0.474 | -0.0628 | No |
| 102 | Slamf1 | 11580 | -0.504 | -0.0576 | No |
| 103 | Kif1b | 11706 | -0.532 | -0.0501 | No |
| 104 | Nmi | 11752 | -0.545 | -0.0358 | No |
| 105 | Ly6e | 11794 | -0.554 | -0.0208 | No |
| 106 | Ptpre | 11937 | -0.606 | -0.0122 | No |
| 107 | Bst2 | 12139 | -0.683 | -0.0060 | No |
| 108 | Ffar2 | 12369 | -0.918 | 0.0058 | No |