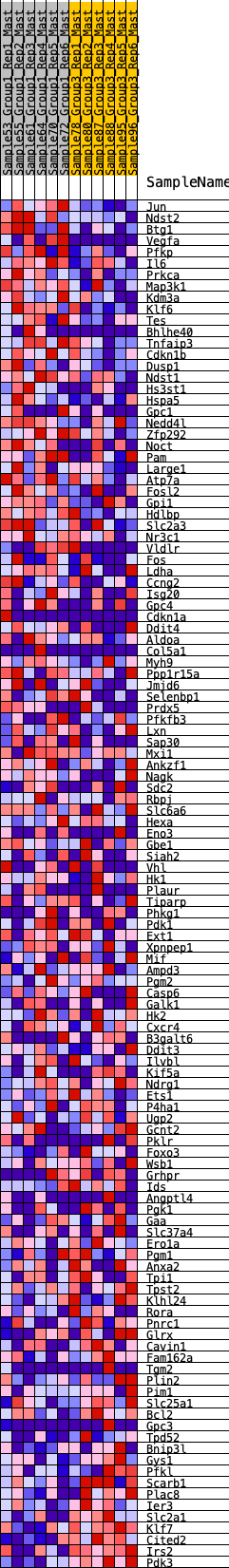
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HYPOXIA |
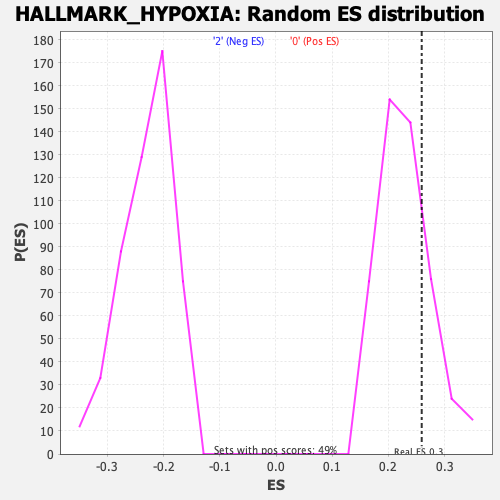
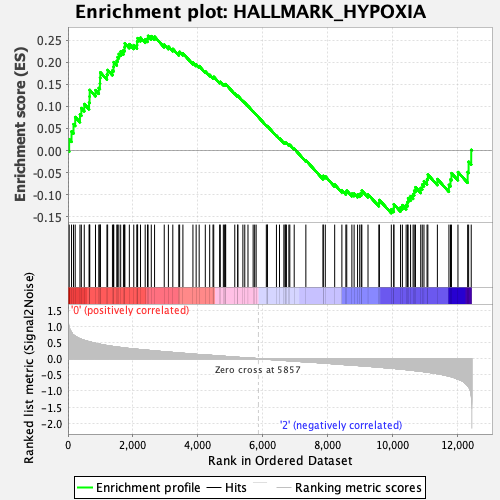
| Enrichment Score (ES) | 0.25939932 |
| Normalized Enrichment Score (NES) | 1.1350783 |
| Nominal p-value | 0.23155738 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jun | 36 | 0.971 | 0.0255 | Yes |
| 2 | Ndst2 | 109 | 0.805 | 0.0432 | Yes |
| 3 | Btg1 | 170 | 0.725 | 0.0595 | Yes |
| 4 | Vegfa | 222 | 0.695 | 0.0757 | Yes |
| 5 | Pfkp | 369 | 0.613 | 0.0818 | Yes |
| 6 | Il6 | 411 | 0.596 | 0.0959 | Yes |
| 7 | Prkca | 501 | 0.563 | 0.1051 | Yes |
| 8 | Map3k1 | 649 | 0.520 | 0.1084 | Yes |
| 9 | Kdm3a | 665 | 0.515 | 0.1222 | Yes |
| 10 | Klf6 | 669 | 0.514 | 0.1370 | Yes |
| 11 | Tes | 847 | 0.473 | 0.1365 | Yes |
| 12 | Bhlhe40 | 949 | 0.451 | 0.1415 | Yes |
| 13 | Tnfaip3 | 983 | 0.444 | 0.1518 | Yes |
| 14 | Cdkn1b | 988 | 0.444 | 0.1645 | Yes |
| 15 | Dusp1 | 996 | 0.442 | 0.1768 | Yes |
| 16 | Ndst1 | 1200 | 0.403 | 0.1721 | Yes |
| 17 | Hs3st1 | 1221 | 0.400 | 0.1822 | Yes |
| 18 | Hspa5 | 1369 | 0.377 | 0.1813 | Yes |
| 19 | Gpc1 | 1403 | 0.372 | 0.1895 | Yes |
| 20 | Nedd4l | 1409 | 0.371 | 0.1999 | Yes |
| 21 | Zfp292 | 1504 | 0.358 | 0.2028 | Yes |
| 22 | Noct | 1529 | 0.355 | 0.2112 | Yes |
| 23 | Pam | 1566 | 0.349 | 0.2185 | Yes |
| 24 | Large1 | 1623 | 0.344 | 0.2240 | Yes |
| 25 | Atp7a | 1708 | 0.332 | 0.2269 | Yes |
| 26 | Fosl2 | 1743 | 0.328 | 0.2337 | Yes |
| 27 | Gpi1 | 1751 | 0.327 | 0.2427 | Yes |
| 28 | Hdlbp | 1890 | 0.311 | 0.2406 | Yes |
| 29 | Slc2a3 | 2026 | 0.299 | 0.2384 | Yes |
| 30 | Nr3c1 | 2125 | 0.288 | 0.2389 | Yes |
| 31 | Vldlr | 2130 | 0.288 | 0.2470 | Yes |
| 32 | Fos | 2145 | 0.286 | 0.2542 | Yes |
| 33 | Ldha | 2234 | 0.278 | 0.2552 | Yes |
| 34 | Ccng2 | 2378 | 0.267 | 0.2514 | Yes |
| 35 | Isg20 | 2449 | 0.259 | 0.2533 | Yes |
| 36 | Gpc4 | 2467 | 0.257 | 0.2594 | Yes |
| 37 | Cdkn1a | 2566 | 0.248 | 0.2587 | No |
| 38 | Ddit4 | 2667 | 0.240 | 0.2576 | No |
| 39 | Aldoa | 2965 | 0.213 | 0.2397 | No |
| 40 | Col5a1 | 3093 | 0.201 | 0.2353 | No |
| 41 | Myh9 | 3229 | 0.190 | 0.2299 | No |
| 42 | Ppp1r15a | 3414 | 0.175 | 0.2201 | No |
| 43 | Jmjd6 | 3439 | 0.173 | 0.2232 | No |
| 44 | Selenbp1 | 3540 | 0.164 | 0.2199 | No |
| 45 | Prdx5 | 3849 | 0.140 | 0.1990 | No |
| 46 | Pfkfb3 | 3948 | 0.131 | 0.1949 | No |
| 47 | Lxn | 4041 | 0.124 | 0.1910 | No |
| 48 | Sap30 | 4230 | 0.113 | 0.1791 | No |
| 49 | Mxi1 | 4365 | 0.103 | 0.1712 | No |
| 50 | Ankzf1 | 4468 | 0.096 | 0.1657 | No |
| 51 | Nagk | 4490 | 0.095 | 0.1668 | No |
| 52 | Sdc2 | 4672 | 0.080 | 0.1545 | No |
| 53 | Rbpj | 4691 | 0.079 | 0.1553 | No |
| 54 | Slc6a6 | 4786 | 0.073 | 0.1498 | No |
| 55 | Hexa | 4818 | 0.070 | 0.1493 | No |
| 56 | Eno3 | 4847 | 0.068 | 0.1490 | No |
| 57 | Gbe1 | 4858 | 0.067 | 0.1502 | No |
| 58 | Siah2 | 5138 | 0.047 | 0.1289 | No |
| 59 | Vhl | 5228 | 0.041 | 0.1229 | No |
| 60 | Hk1 | 5233 | 0.041 | 0.1238 | No |
| 61 | Plaur | 5388 | 0.029 | 0.1121 | No |
| 62 | Tiparp | 5446 | 0.025 | 0.1082 | No |
| 63 | Phkg1 | 5545 | 0.019 | 0.1008 | No |
| 64 | Pdk1 | 5704 | 0.010 | 0.0883 | No |
| 65 | Ext1 | 5751 | 0.007 | 0.0848 | No |
| 66 | Xpnpep1 | 5796 | 0.004 | 0.0813 | No |
| 67 | Mif | 6110 | -0.018 | 0.0564 | No |
| 68 | Ampd3 | 6139 | -0.020 | 0.0548 | No |
| 69 | Pgm2 | 6142 | -0.021 | 0.0552 | No |
| 70 | Casp6 | 6422 | -0.039 | 0.0337 | No |
| 71 | Galk1 | 6516 | -0.045 | 0.0275 | No |
| 72 | Hk2 | 6651 | -0.055 | 0.0182 | No |
| 73 | Cxcr4 | 6686 | -0.057 | 0.0171 | No |
| 74 | B3galt6 | 6705 | -0.058 | 0.0174 | No |
| 75 | Ddit3 | 6729 | -0.060 | 0.0172 | No |
| 76 | Ilvbl | 6805 | -0.065 | 0.0130 | No |
| 77 | Kif5a | 6834 | -0.067 | 0.0127 | No |
| 78 | Ndrg1 | 6969 | -0.075 | 0.0040 | No |
| 79 | Ets1 | 7328 | -0.099 | -0.0221 | No |
| 80 | P4ha1 | 7860 | -0.135 | -0.0612 | No |
| 81 | Ugp2 | 7862 | -0.135 | -0.0574 | No |
| 82 | Gcnt2 | 7927 | -0.140 | -0.0585 | No |
| 83 | Pklr | 8216 | -0.161 | -0.0771 | No |
| 84 | Foxo3 | 8438 | -0.178 | -0.0898 | No |
| 85 | Wsb1 | 8561 | -0.187 | -0.0943 | No |
| 86 | Grhpr | 8588 | -0.190 | -0.0908 | No |
| 87 | Ids | 8746 | -0.200 | -0.0977 | No |
| 88 | Angptl4 | 8815 | -0.205 | -0.0973 | No |
| 89 | Pgk1 | 8921 | -0.213 | -0.0996 | No |
| 90 | Gaa | 8987 | -0.216 | -0.0985 | No |
| 91 | Slc37a4 | 9035 | -0.221 | -0.0959 | No |
| 92 | Ero1a | 9053 | -0.222 | -0.0908 | No |
| 93 | Pgm1 | 9243 | -0.236 | -0.0992 | No |
| 94 | Anxa2 | 9578 | -0.265 | -0.1186 | No |
| 95 | Tpi1 | 9594 | -0.266 | -0.1120 | No |
| 96 | Tpst2 | 9961 | -0.297 | -0.1330 | No |
| 97 | Klhl24 | 10038 | -0.303 | -0.1303 | No |
| 98 | Rora | 10039 | -0.303 | -0.1215 | No |
| 99 | Pnrc1 | 10245 | -0.323 | -0.1287 | No |
| 100 | Glrx | 10302 | -0.327 | -0.1237 | No |
| 101 | Cavin1 | 10418 | -0.339 | -0.1231 | No |
| 102 | Fam162a | 10462 | -0.343 | -0.1165 | No |
| 103 | Tgm2 | 10478 | -0.345 | -0.1076 | No |
| 104 | Plin2 | 10554 | -0.354 | -0.1034 | No |
| 105 | Pim1 | 10632 | -0.364 | -0.0990 | No |
| 106 | Slc25a1 | 10666 | -0.368 | -0.0909 | No |
| 107 | Bcl2 | 10705 | -0.372 | -0.0831 | No |
| 108 | Gpc3 | 10867 | -0.389 | -0.0848 | No |
| 109 | Tpd52 | 10914 | -0.396 | -0.0770 | No |
| 110 | Bnip3l | 10967 | -0.403 | -0.0694 | No |
| 111 | Gys1 | 11062 | -0.415 | -0.0649 | No |
| 112 | Pfkl | 11087 | -0.419 | -0.0546 | No |
| 113 | Scarb1 | 11380 | -0.466 | -0.0646 | No |
| 114 | Plac8 | 11736 | -0.542 | -0.0776 | No |
| 115 | Ier3 | 11784 | -0.551 | -0.0653 | No |
| 116 | Slc2a1 | 11814 | -0.561 | -0.0513 | No |
| 117 | Klf7 | 12014 | -0.637 | -0.0488 | No |
| 118 | Cited2 | 12313 | -0.830 | -0.0487 | No |
| 119 | Irs2 | 12343 | -0.872 | -0.0256 | No |
| 120 | Pdk3 | 12423 | -1.143 | 0.0015 | No |