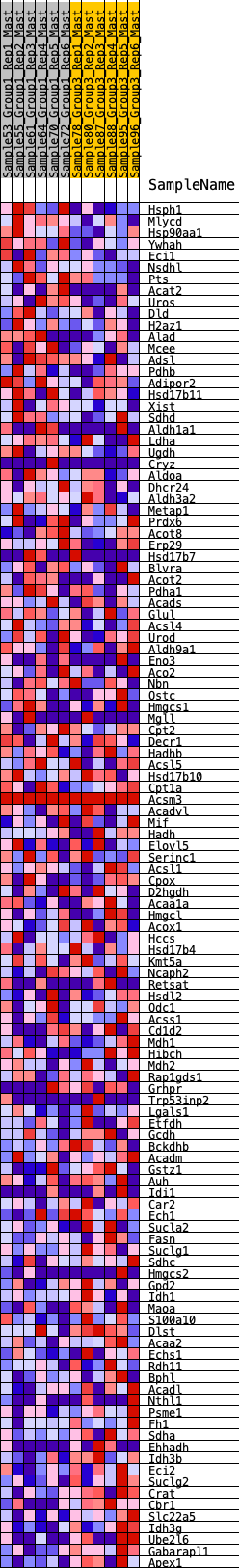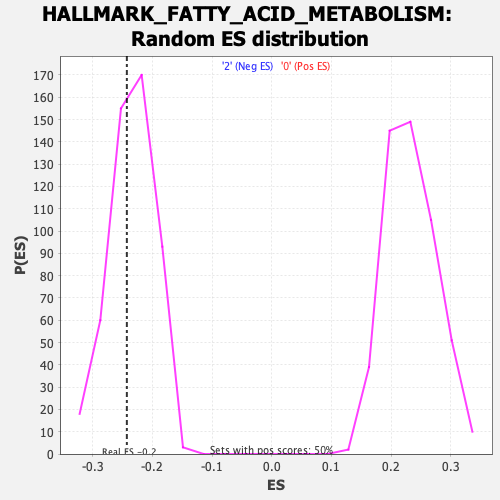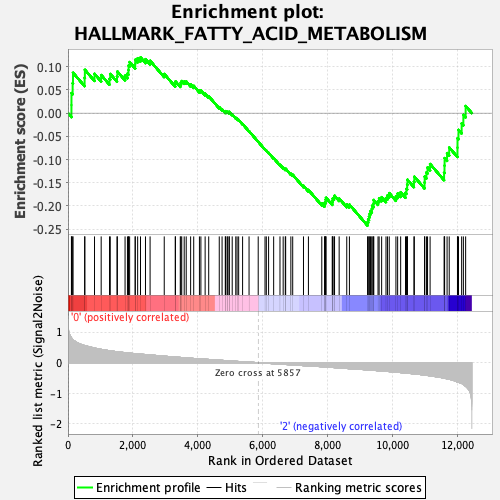
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2425523 |
| Normalized Enrichment Score (NES) | -1.0353084 |
| Nominal p-value | 0.3987976 |
| FDR q-value | 0.8729905 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsph1 | 106 | 0.810 | 0.0173 | No |
| 2 | Mlycd | 107 | 0.810 | 0.0432 | No |
| 3 | Hsp90aa1 | 143 | 0.749 | 0.0644 | No |
| 4 | Ywhah | 154 | 0.737 | 0.0872 | No |
| 5 | Eci1 | 512 | 0.561 | 0.0762 | No |
| 6 | Nsdhl | 520 | 0.559 | 0.0935 | No |
| 7 | Pts | 817 | 0.479 | 0.0848 | No |
| 8 | Acat2 | 1022 | 0.434 | 0.0822 | No |
| 9 | Uros | 1279 | 0.391 | 0.0739 | No |
| 10 | Dld | 1302 | 0.387 | 0.0845 | No |
| 11 | H2az1 | 1510 | 0.358 | 0.0792 | No |
| 12 | Alad | 1521 | 0.356 | 0.0898 | No |
| 13 | Mcee | 1758 | 0.326 | 0.0811 | No |
| 14 | Adsl | 1832 | 0.317 | 0.0853 | No |
| 15 | Pdhb | 1863 | 0.314 | 0.0929 | No |
| 16 | Adipor2 | 1867 | 0.314 | 0.1027 | No |
| 17 | Hsd17b11 | 1899 | 0.310 | 0.1101 | No |
| 18 | Xist | 2070 | 0.294 | 0.1058 | No |
| 19 | Sdhd | 2071 | 0.294 | 0.1152 | No |
| 20 | Aldh1a1 | 2146 | 0.286 | 0.1184 | No |
| 21 | Ldha | 2234 | 0.278 | 0.1202 | No |
| 22 | Ugdh | 2387 | 0.265 | 0.1164 | No |
| 23 | Cryz | 2529 | 0.251 | 0.1130 | No |
| 24 | Aldoa | 2965 | 0.213 | 0.0845 | No |
| 25 | Dhcr24 | 3304 | 0.184 | 0.0630 | No |
| 26 | Aldh3a2 | 3311 | 0.183 | 0.0683 | No |
| 27 | Metap1 | 3459 | 0.171 | 0.0619 | No |
| 28 | Prdx6 | 3477 | 0.170 | 0.0660 | No |
| 29 | Acot8 | 3504 | 0.168 | 0.0692 | No |
| 30 | Erp29 | 3580 | 0.161 | 0.0683 | No |
| 31 | Hsd17b7 | 3642 | 0.157 | 0.0684 | No |
| 32 | Blvra | 3778 | 0.145 | 0.0621 | No |
| 33 | Acot2 | 3873 | 0.137 | 0.0588 | No |
| 34 | Pdha1 | 4049 | 0.123 | 0.0486 | No |
| 35 | Acads | 4094 | 0.120 | 0.0489 | No |
| 36 | Glul | 4225 | 0.113 | 0.0419 | No |
| 37 | Acsl4 | 4335 | 0.105 | 0.0364 | No |
| 38 | Urod | 4660 | 0.081 | 0.0128 | No |
| 39 | Aldh9a1 | 4747 | 0.075 | 0.0082 | No |
| 40 | Eno3 | 4847 | 0.068 | 0.0023 | No |
| 41 | Aco2 | 4851 | 0.068 | 0.0043 | No |
| 42 | Nbn | 4900 | 0.063 | 0.0024 | No |
| 43 | Ostc | 4919 | 0.062 | 0.0029 | No |
| 44 | Hmgcs1 | 4965 | 0.058 | 0.0011 | No |
| 45 | Mgll | 4968 | 0.058 | 0.0028 | No |
| 46 | Cpt2 | 5059 | 0.053 | -0.0028 | No |
| 47 | Decr1 | 5169 | 0.045 | -0.0102 | No |
| 48 | Hadhb | 5209 | 0.042 | -0.0120 | No |
| 49 | Acsl5 | 5258 | 0.039 | -0.0147 | No |
| 50 | Hsd17b10 | 5379 | 0.030 | -0.0235 | No |
| 51 | Cpt1a | 5578 | 0.017 | -0.0390 | No |
| 52 | Acsm3 | 5860 | 0.000 | -0.0618 | No |
| 53 | Acadvl | 6070 | -0.016 | -0.0782 | No |
| 54 | Mif | 6110 | -0.018 | -0.0808 | No |
| 55 | Hadh | 6180 | -0.024 | -0.0856 | No |
| 56 | Elovl5 | 6336 | -0.033 | -0.0971 | No |
| 57 | Serinc1 | 6531 | -0.046 | -0.1114 | No |
| 58 | Acsl1 | 6629 | -0.053 | -0.1176 | No |
| 59 | Cpox | 6694 | -0.057 | -0.1209 | No |
| 60 | D2hgdh | 6706 | -0.058 | -0.1200 | No |
| 61 | Acaa1a | 6866 | -0.069 | -0.1307 | No |
| 62 | Hmgcl | 6923 | -0.072 | -0.1329 | No |
| 63 | Acox1 | 7255 | -0.096 | -0.1567 | No |
| 64 | Hccs | 7407 | -0.104 | -0.1656 | No |
| 65 | Hsd17b4 | 7819 | -0.132 | -0.1947 | No |
| 66 | Kmt5a | 7905 | -0.139 | -0.1972 | No |
| 67 | Ncaph2 | 7910 | -0.139 | -0.1931 | No |
| 68 | Retsat | 7936 | -0.141 | -0.1906 | No |
| 69 | Hsdl2 | 7938 | -0.141 | -0.1862 | No |
| 70 | Odc1 | 7949 | -0.142 | -0.1825 | No |
| 71 | Acss1 | 8142 | -0.156 | -0.1931 | No |
| 72 | Cd1d2 | 8147 | -0.156 | -0.1884 | No |
| 73 | Mdh1 | 8157 | -0.157 | -0.1841 | No |
| 74 | Hibch | 8196 | -0.160 | -0.1821 | No |
| 75 | Mdh2 | 8212 | -0.161 | -0.1781 | No |
| 76 | Rap1gds1 | 8354 | -0.171 | -0.1841 | No |
| 77 | Grhpr | 8588 | -0.190 | -0.1969 | No |
| 78 | Trp53inp2 | 8670 | -0.195 | -0.1973 | No |
| 79 | Lgals1 | 9229 | -0.235 | -0.2350 | Yes |
| 80 | Etfdh | 9248 | -0.237 | -0.2289 | Yes |
| 81 | Gcdh | 9274 | -0.239 | -0.2233 | Yes |
| 82 | Bckdhb | 9296 | -0.240 | -0.2173 | Yes |
| 83 | Acadm | 9319 | -0.243 | -0.2113 | Yes |
| 84 | Gstz1 | 9361 | -0.246 | -0.2067 | Yes |
| 85 | Auh | 9370 | -0.246 | -0.1995 | Yes |
| 86 | Idi1 | 9415 | -0.250 | -0.1951 | Yes |
| 87 | Car2 | 9419 | -0.251 | -0.1873 | Yes |
| 88 | Ech1 | 9552 | -0.263 | -0.1896 | Yes |
| 89 | Sucla2 | 9585 | -0.265 | -0.1837 | Yes |
| 90 | Fasn | 9663 | -0.270 | -0.1813 | Yes |
| 91 | Suclg1 | 9797 | -0.282 | -0.1831 | Yes |
| 92 | Sdhc | 9842 | -0.286 | -0.1775 | Yes |
| 93 | Hmgcs2 | 9897 | -0.291 | -0.1725 | Yes |
| 94 | Gpd2 | 10103 | -0.308 | -0.1793 | Yes |
| 95 | Idh1 | 10156 | -0.314 | -0.1734 | Yes |
| 96 | Maoa | 10249 | -0.324 | -0.1705 | Yes |
| 97 | S100a10 | 10398 | -0.337 | -0.1718 | Yes |
| 98 | Dlst | 10428 | -0.340 | -0.1632 | Yes |
| 99 | Acaa2 | 10450 | -0.342 | -0.1540 | Yes |
| 100 | Echs1 | 10459 | -0.343 | -0.1436 | Yes |
| 101 | Rdh11 | 10658 | -0.367 | -0.1479 | Yes |
| 102 | Bphl | 10670 | -0.368 | -0.1370 | Yes |
| 103 | Acadl | 10985 | -0.405 | -0.1495 | Yes |
| 104 | Nthl1 | 10990 | -0.406 | -0.1369 | Yes |
| 105 | Psme1 | 11043 | -0.413 | -0.1279 | Yes |
| 106 | Fh1 | 11077 | -0.417 | -0.1172 | Yes |
| 107 | Sdha | 11156 | -0.430 | -0.1097 | Yes |
| 108 | Ehhadh | 11587 | -0.505 | -0.1285 | Yes |
| 109 | Idh3b | 11600 | -0.509 | -0.1131 | Yes |
| 110 | Eci2 | 11603 | -0.509 | -0.0970 | Yes |
| 111 | Suclg2 | 11679 | -0.527 | -0.0862 | Yes |
| 112 | Crat | 11745 | -0.544 | -0.0741 | Yes |
| 113 | Cbr1 | 11999 | -0.630 | -0.0744 | Yes |
| 114 | Slc22a5 | 12001 | -0.632 | -0.0543 | Yes |
| 115 | Idh3g | 12031 | -0.641 | -0.0361 | Yes |
| 116 | Ube2l6 | 12128 | -0.678 | -0.0222 | Yes |
| 117 | Gabarapl1 | 12181 | -0.709 | -0.0037 | Yes |
| 118 | Apex1 | 12251 | -0.772 | 0.0154 | Yes |