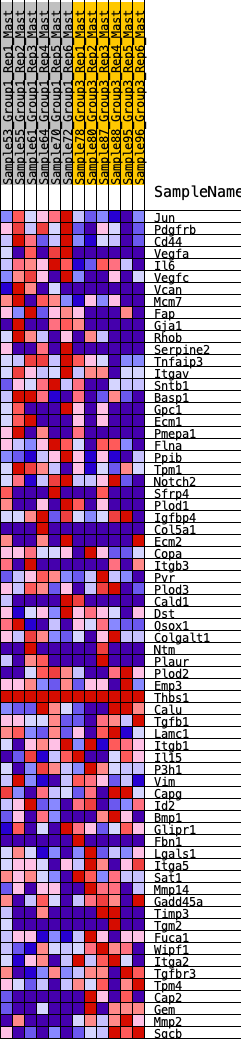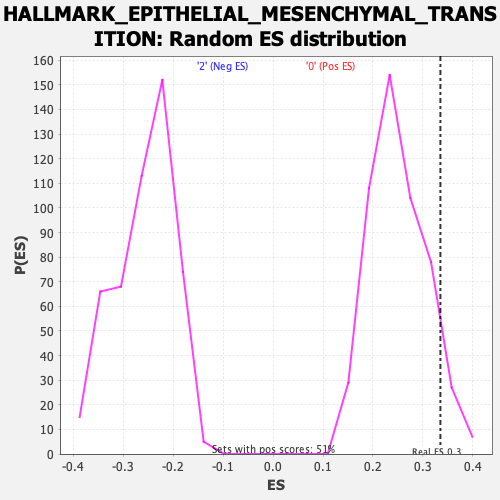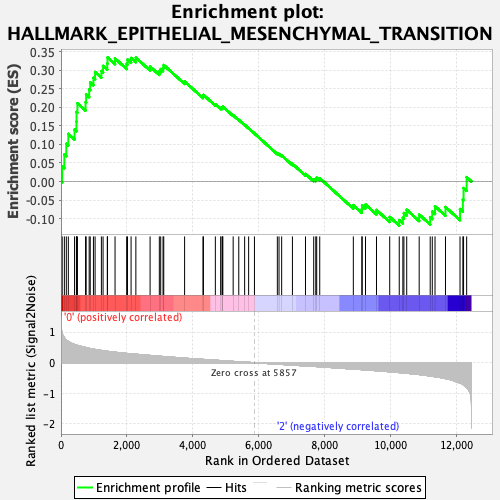
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.3351547 |
| Normalized Enrichment Score (NES) | 1.3330848 |
| Nominal p-value | 0.0729783 |
| FDR q-value | 0.4221589 |
| FWER p-Value | 0.762 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jun | 36 | 0.971 | 0.0412 | Yes |
| 2 | Pdgfrb | 102 | 0.821 | 0.0732 | Yes |
| 3 | Cd44 | 161 | 0.734 | 0.1019 | Yes |
| 4 | Vegfa | 222 | 0.695 | 0.1286 | Yes |
| 5 | Il6 | 411 | 0.596 | 0.1404 | Yes |
| 6 | Vegfc | 468 | 0.574 | 0.1620 | Yes |
| 7 | Vcan | 471 | 0.573 | 0.1878 | Yes |
| 8 | Mcm7 | 498 | 0.565 | 0.2114 | Yes |
| 9 | Fap | 744 | 0.498 | 0.2142 | Yes |
| 10 | Gja1 | 762 | 0.492 | 0.2351 | Yes |
| 11 | Rhob | 854 | 0.471 | 0.2492 | Yes |
| 12 | Serpine2 | 886 | 0.463 | 0.2677 | Yes |
| 13 | Tnfaip3 | 983 | 0.444 | 0.2801 | Yes |
| 14 | Itgav | 1033 | 0.432 | 0.2957 | Yes |
| 15 | Sntb1 | 1224 | 0.399 | 0.2985 | Yes |
| 16 | Basp1 | 1275 | 0.391 | 0.3122 | Yes |
| 17 | Gpc1 | 1403 | 0.372 | 0.3188 | Yes |
| 18 | Ecm1 | 1410 | 0.371 | 0.3352 | Yes |
| 19 | Pmepa1 | 1638 | 0.340 | 0.3323 | No |
| 20 | Flna | 1990 | 0.302 | 0.3176 | No |
| 21 | Ppib | 2016 | 0.300 | 0.3292 | No |
| 22 | Tpm1 | 2124 | 0.288 | 0.3336 | No |
| 23 | Notch2 | 2269 | 0.275 | 0.3345 | No |
| 24 | Sfrp4 | 2700 | 0.236 | 0.3104 | No |
| 25 | Plod1 | 2980 | 0.211 | 0.2975 | No |
| 26 | Igfbp4 | 3022 | 0.208 | 0.3036 | No |
| 27 | Col5a1 | 3093 | 0.201 | 0.3071 | No |
| 28 | Ecm2 | 3114 | 0.200 | 0.3145 | No |
| 29 | Copa | 3746 | 0.149 | 0.2703 | No |
| 30 | Itgb3 | 4303 | 0.107 | 0.2302 | No |
| 31 | Pvr | 4317 | 0.107 | 0.2340 | No |
| 32 | Plod3 | 4678 | 0.080 | 0.2085 | No |
| 33 | Cald1 | 4839 | 0.069 | 0.1987 | No |
| 34 | Dst | 4880 | 0.065 | 0.1984 | No |
| 35 | Qsox1 | 4899 | 0.063 | 0.1998 | No |
| 36 | Colgalt1 | 4902 | 0.063 | 0.2025 | No |
| 37 | Ntm | 5218 | 0.041 | 0.1789 | No |
| 38 | Plaur | 5388 | 0.029 | 0.1666 | No |
| 39 | Plod2 | 5566 | 0.018 | 0.1531 | No |
| 40 | Emp3 | 5687 | 0.011 | 0.1439 | No |
| 41 | Thbs1 | 5863 | 0.000 | 0.1298 | No |
| 42 | Calu | 6557 | -0.048 | 0.0759 | No |
| 43 | Tgfb1 | 6608 | -0.052 | 0.0742 | No |
| 44 | Lamc1 | 6688 | -0.057 | 0.0705 | No |
| 45 | Itgb1 | 7014 | -0.078 | 0.0477 | No |
| 46 | Il15 | 7410 | -0.104 | 0.0205 | No |
| 47 | P3h1 | 7659 | -0.120 | 0.0059 | No |
| 48 | Vim | 7722 | -0.125 | 0.0066 | No |
| 49 | Capg | 7750 | -0.127 | 0.0102 | No |
| 50 | Id2 | 7842 | -0.133 | 0.0089 | No |
| 51 | Bmp1 | 8860 | -0.208 | -0.0639 | No |
| 52 | Glipr1 | 9119 | -0.227 | -0.0744 | No |
| 53 | Fbn1 | 9128 | -0.227 | -0.0647 | No |
| 54 | Lgals1 | 9229 | -0.235 | -0.0621 | No |
| 55 | Itga5 | 9562 | -0.264 | -0.0770 | No |
| 56 | Sat1 | 9963 | -0.297 | -0.0958 | No |
| 57 | Mmp14 | 10252 | -0.324 | -0.1044 | No |
| 58 | Gadd45a | 10361 | -0.333 | -0.0980 | No |
| 59 | Timp3 | 10393 | -0.337 | -0.0852 | No |
| 60 | Tgm2 | 10478 | -0.345 | -0.0764 | No |
| 61 | Fuca1 | 10856 | -0.388 | -0.0892 | No |
| 62 | Wipf1 | 11190 | -0.435 | -0.0964 | No |
| 63 | Itga2 | 11253 | -0.445 | -0.0812 | No |
| 64 | Tgfbr3 | 11336 | -0.460 | -0.0670 | No |
| 65 | Tpm4 | 11653 | -0.520 | -0.0689 | No |
| 66 | Cap2 | 12094 | -0.662 | -0.0744 | No |
| 67 | Gem | 12180 | -0.708 | -0.0492 | No |
| 68 | Mmp2 | 12201 | -0.729 | -0.0177 | No |
| 69 | Sgcb | 12295 | -0.815 | 0.0118 | No |