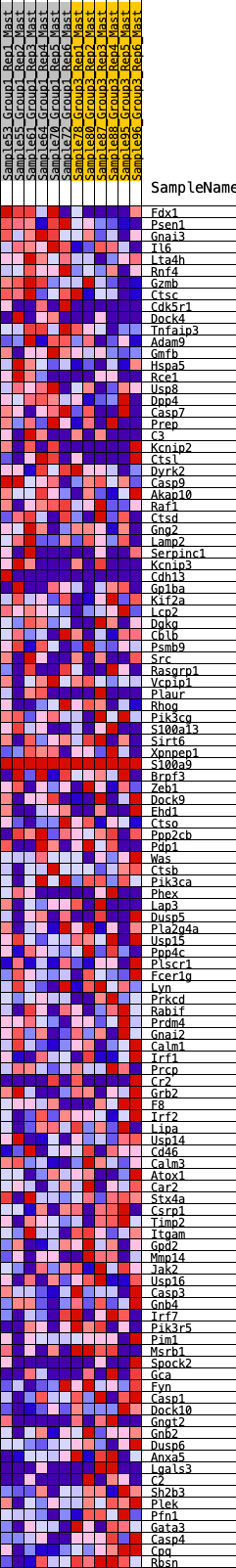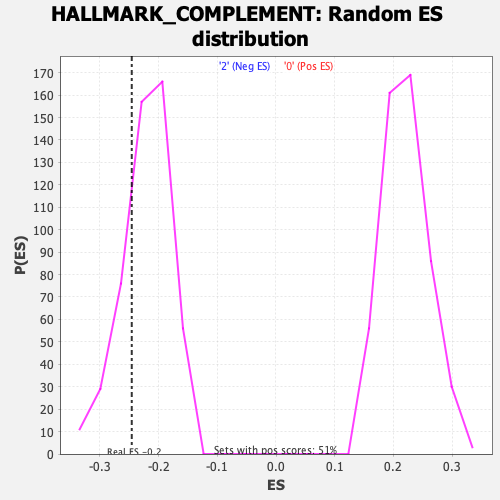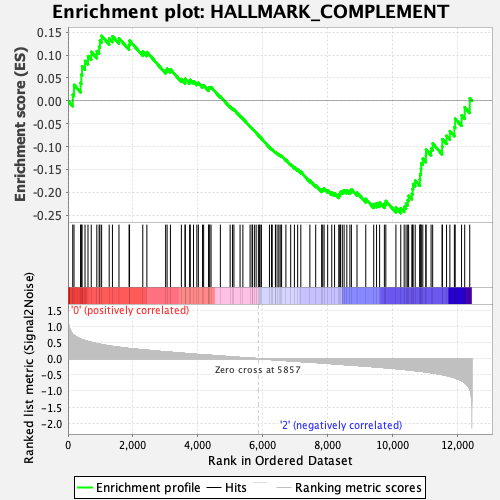
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.24563324 |
| Normalized Enrichment Score (NES) | -1.1085669 |
| Nominal p-value | 0.24242425 |
| FDR q-value | 0.7179245 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdx1 | 146 | 0.742 | 0.0133 | No |
| 2 | Psen1 | 185 | 0.719 | 0.0347 | No |
| 3 | Gnai3 | 387 | 0.605 | 0.0389 | No |
| 4 | Il6 | 411 | 0.596 | 0.0572 | No |
| 5 | Lta4h | 434 | 0.586 | 0.0753 | No |
| 6 | Rnf4 | 524 | 0.558 | 0.0871 | No |
| 7 | Gzmb | 616 | 0.531 | 0.0977 | No |
| 8 | Ctsc | 716 | 0.505 | 0.1068 | No |
| 9 | Cdk5r1 | 888 | 0.463 | 0.1087 | No |
| 10 | Dock4 | 959 | 0.449 | 0.1182 | No |
| 11 | Tnfaip3 | 983 | 0.444 | 0.1314 | No |
| 12 | Adam9 | 1036 | 0.431 | 0.1418 | No |
| 13 | Gmfb | 1269 | 0.393 | 0.1364 | No |
| 14 | Hspa5 | 1369 | 0.377 | 0.1411 | No |
| 15 | Rce1 | 1570 | 0.349 | 0.1367 | No |
| 16 | Usp8 | 1882 | 0.312 | 0.1221 | No |
| 17 | Dpp4 | 1896 | 0.311 | 0.1316 | No |
| 18 | Casp7 | 2304 | 0.272 | 0.1078 | No |
| 19 | Prep | 2430 | 0.260 | 0.1065 | No |
| 20 | C3 | 3008 | 0.209 | 0.0668 | No |
| 21 | Kcnip2 | 3053 | 0.205 | 0.0702 | No |
| 22 | Ctsl | 3156 | 0.197 | 0.0686 | No |
| 23 | Dyrk2 | 3493 | 0.168 | 0.0470 | No |
| 24 | Casp9 | 3607 | 0.159 | 0.0433 | No |
| 25 | Akap10 | 3612 | 0.159 | 0.0483 | No |
| 26 | Raf1 | 3747 | 0.149 | 0.0425 | No |
| 27 | Ctsd | 3767 | 0.147 | 0.0459 | No |
| 28 | Gng2 | 3866 | 0.138 | 0.0427 | No |
| 29 | Lamp2 | 3970 | 0.129 | 0.0387 | No |
| 30 | Serpinc1 | 4016 | 0.126 | 0.0393 | No |
| 31 | Kcnip3 | 4145 | 0.116 | 0.0329 | No |
| 32 | Cdh13 | 4179 | 0.114 | 0.0341 | No |
| 33 | Gp1ba | 4334 | 0.105 | 0.0252 | No |
| 34 | Kif2a | 4338 | 0.105 | 0.0285 | No |
| 35 | Lcp2 | 4368 | 0.103 | 0.0296 | No |
| 36 | Dgkg | 4407 | 0.100 | 0.0299 | No |
| 37 | Cblb | 4694 | 0.078 | 0.0094 | No |
| 38 | Psmb9 | 4995 | 0.056 | -0.0130 | No |
| 39 | Src | 5067 | 0.052 | -0.0170 | No |
| 40 | Rasgrp1 | 5112 | 0.049 | -0.0189 | No |
| 41 | Vcpip1 | 5301 | 0.035 | -0.0330 | No |
| 42 | Plaur | 5388 | 0.029 | -0.0390 | No |
| 43 | Rhog | 5610 | 0.015 | -0.0564 | No |
| 44 | Pik3cg | 5673 | 0.012 | -0.0610 | No |
| 45 | S100a13 | 5679 | 0.012 | -0.0610 | No |
| 46 | Sirt6 | 5742 | 0.008 | -0.0658 | No |
| 47 | Xpnpep1 | 5796 | 0.004 | -0.0699 | No |
| 48 | S100a9 | 5865 | 0.000 | -0.0755 | No |
| 49 | Brpf3 | 5896 | -0.002 | -0.0778 | No |
| 50 | Zeb1 | 5918 | -0.003 | -0.0794 | No |
| 51 | Dock9 | 5959 | -0.008 | -0.0824 | No |
| 52 | Ehd1 | 6207 | -0.025 | -0.1016 | No |
| 53 | Ctso | 6269 | -0.029 | -0.1055 | No |
| 54 | Ppp2cb | 6292 | -0.031 | -0.1063 | No |
| 55 | Pdp1 | 6390 | -0.037 | -0.1129 | No |
| 56 | Was | 6405 | -0.038 | -0.1127 | No |
| 57 | Ctsb | 6468 | -0.042 | -0.1163 | No |
| 58 | Pik3ca | 6502 | -0.044 | -0.1175 | No |
| 59 | Phex | 6556 | -0.048 | -0.1202 | No |
| 60 | Lap3 | 6581 | -0.050 | -0.1204 | No |
| 61 | Dusp5 | 6713 | -0.059 | -0.1291 | No |
| 62 | Pla2g4a | 6861 | -0.068 | -0.1387 | No |
| 63 | Usp15 | 6975 | -0.076 | -0.1453 | No |
| 64 | Ppp4c | 7075 | -0.083 | -0.1505 | No |
| 65 | Plscr1 | 7173 | -0.090 | -0.1553 | No |
| 66 | Fcer1g | 7452 | -0.107 | -0.1742 | No |
| 67 | Lyn | 7631 | -0.119 | -0.1846 | No |
| 68 | Prkcd | 7814 | -0.131 | -0.1949 | No |
| 69 | Rabif | 7848 | -0.134 | -0.1930 | No |
| 70 | Prdm4 | 7890 | -0.137 | -0.1917 | No |
| 71 | Gnai2 | 8000 | -0.145 | -0.1956 | No |
| 72 | Calm1 | 8126 | -0.154 | -0.2005 | No |
| 73 | Irf1 | 8213 | -0.161 | -0.2020 | No |
| 74 | Prcp | 8342 | -0.170 | -0.2066 | No |
| 75 | Cr2 | 8372 | -0.173 | -0.2031 | No |
| 76 | Grb2 | 8397 | -0.174 | -0.1992 | No |
| 77 | F8 | 8451 | -0.179 | -0.1974 | No |
| 78 | Irf2 | 8507 | -0.183 | -0.1956 | No |
| 79 | Lipa | 8590 | -0.190 | -0.1958 | No |
| 80 | Usp14 | 8685 | -0.195 | -0.1968 | No |
| 81 | Cd46 | 8731 | -0.199 | -0.1937 | No |
| 82 | Calm3 | 8904 | -0.211 | -0.2005 | No |
| 83 | Atox1 | 9174 | -0.232 | -0.2145 | No |
| 84 | Car2 | 9419 | -0.251 | -0.2258 | No |
| 85 | Stx4a | 9506 | -0.258 | -0.2240 | No |
| 86 | Csrp1 | 9601 | -0.266 | -0.2226 | No |
| 87 | Timp2 | 9747 | -0.277 | -0.2249 | No |
| 88 | Itgam | 9792 | -0.281 | -0.2189 | No |
| 89 | Gpd2 | 10103 | -0.308 | -0.2336 | No |
| 90 | Mmp14 | 10252 | -0.324 | -0.2346 | Yes |
| 91 | Jak2 | 10357 | -0.333 | -0.2318 | Yes |
| 92 | Usp16 | 10415 | -0.339 | -0.2249 | Yes |
| 93 | Casp3 | 10466 | -0.344 | -0.2173 | Yes |
| 94 | Gnb4 | 10494 | -0.347 | -0.2077 | Yes |
| 95 | Irf7 | 10594 | -0.359 | -0.2035 | Yes |
| 96 | Pik3r5 | 10616 | -0.362 | -0.1930 | Yes |
| 97 | Pim1 | 10632 | -0.364 | -0.1818 | Yes |
| 98 | Msrb1 | 10697 | -0.370 | -0.1744 | Yes |
| 99 | Spock2 | 10837 | -0.386 | -0.1726 | Yes |
| 100 | Gca | 10847 | -0.387 | -0.1602 | Yes |
| 101 | Fyn | 10873 | -0.390 | -0.1490 | Yes |
| 102 | Casp1 | 10882 | -0.391 | -0.1364 | Yes |
| 103 | Dock10 | 10931 | -0.398 | -0.1267 | Yes |
| 104 | Gngt2 | 11026 | -0.410 | -0.1204 | Yes |
| 105 | Gnb2 | 11030 | -0.411 | -0.1067 | Yes |
| 106 | Dusp6 | 11186 | -0.434 | -0.1046 | Yes |
| 107 | Anxa5 | 11235 | -0.443 | -0.0934 | Yes |
| 108 | Lgals3 | 11526 | -0.492 | -0.1002 | Yes |
| 109 | C2 | 11534 | -0.494 | -0.0841 | Yes |
| 110 | Sh2b3 | 11663 | -0.523 | -0.0767 | Yes |
| 111 | Plek | 11765 | -0.546 | -0.0663 | Yes |
| 112 | Pfn1 | 11904 | -0.590 | -0.0575 | Yes |
| 113 | Gata3 | 11929 | -0.603 | -0.0390 | Yes |
| 114 | Casp4 | 12125 | -0.675 | -0.0319 | Yes |
| 115 | Cpq | 12222 | -0.748 | -0.0143 | Yes |
| 116 | Rbsn | 12377 | -0.941 | 0.0052 | Yes |